













| General Information: |
|
| Name(s) found: |
gi|213053374
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Salmonella enterica subsp. enterica serovar Typhi str. E00-7866 |
| Length: | 847 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVRLAIIG NGMVGHRFIE DLLDKSDASL FDITVFCEEP RKAYDRVHLS SYFSHHTAEE 60
61 LSLVREGFYE KHGVKVLVGE RAITINRQEK VIHSSAGRTV YYDKLIMATG SYPWIPPIKG 120
121 SETQDCFVYR TIEDLNAIEA CARRSKRGAV VGGGLLGLEA AGALKNLGVE THVIEFAPML 180
181 MAEQLDQMGG EQLRRKIESM GVRVHTSKNT QEIVQQGNEA RKTMRFADGS ELEVDFIVFS 240
241 TGIRPRDKLA TQCGLAVAQR GGIVINDTCQ TSDPDIYAIG ECASWNNRVY GLVAPGYKMA 300
301 QVAVDHILGS ENAFEGADLS AKLKLLGVDV GGIGDAHGRT PGARSYVYLD ESKEVYKRLI 360
361 VSEDNKTLLG AVLVGDTSDY GNLLQLVLNA IELPENPDSL ILPSHASSGK PAIGVDKLPD 420
421 SAQICSCFDV TKGDLIAAIN KGCHTVAALK AETKAGTGCG GCIPLVTQVL NAELAKQGIE 480
481 VNNNLCEHFA YSRQELFHLI RVEGIKTFEE LLTKHGKGYG CEVCKPTVGS LLASCWNEYI 540
541 LKPQHTPLQD TNDNFLANIQ KDGTYSVIPR SAGGEITPEG LVAVGRIARE FNLYTKITGS 600
601 QRIGLFGAQK DDLPEIWRQL IEAGFETGHA YAKALRMAKT CVGSTWCRYG VGDSVGFGVE 660
661 LENRYKGIRT PHKMKFGVSG CTRECAEAQG KDVGIIATEK GWNLYVCGNG GMKPRHADLL 720
721 AADLDRDTLI KYLDRFMMFY IRTADKLTRT APWLDNLEGG IEYLKSVIID DKLGLNEHLE 780
781 EEMARLRAAV VCEWTETVNT PAAQVRFKHF INSDKRDPNV QVVPEREQHR PATPYERIPV 840
841 TLVEENA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
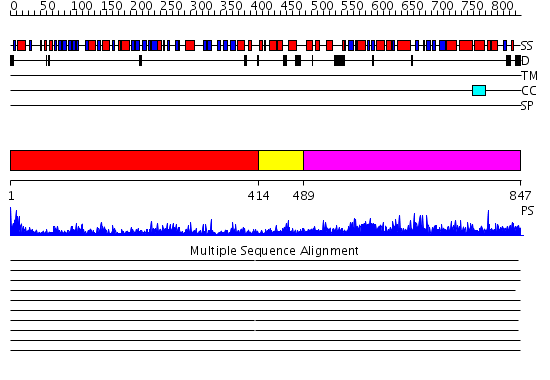
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..413] | 79.0 | Dihydrolipoamide dehydrogenase | |
| 2 | View Details | [414..488] | 0.968 | Crystal structure of L-proline dehydrogenase from P.horikoshii | |
| 3 | View Details | [489..847] | 68.69897 | Structure of spinach nitrite reductase |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)