













| General Information: |
|
| Name(s) found: |
gi|190405000
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae RM11-1a |
| Length: | 889 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSISETPHNK SQGLQKAAGR PKIVVPEGSP SRNSDSGSFT IEGDTSLNDD LLSISGSVTP 60
61 RARRSSRLSL DSITPRRSFD SRTLSVANSR SFGFENETHS GSMDFSPLGN NSIYEIVMNT 120
121 RRKNWLNYPT VADIPQVSLS KNDLDDHWKT HVIEYVKNIK SDYQIFQSTN NIRNMNQMEQ 180
181 LKELREGENM HEESFEANLR QGDAELINSI PDFYFSDKFQ LDNPRTFHKV LDAIDLFLTK 240
241 LDMKRQAERD EAFSELRDRL NDFLDIVETL LVTEISKSSH KFFHALSEVD NIQKRALDTM 300
301 SELKELAQNI KTIDAENIRK KISHLEMIFK RKNVEKLEQG LLQAKLVLNK TDECKSMYEE 360
361 NKLDNCLELI KSIDYLIKGD DSINEDVQSW TRCWPYKLSN LRTIPALSAT REFLTNMKIE 420
421 IGGKFSLQLS ILLIDDLRSF CKSIKPKETL HRIQTGSNDK KQTIFTDNFS SKITELIVRL 480
481 NRCEELTSAF DLYREKSITE LKSIIKIYLP TENAHADNNH DEKHLNNGST SGSKLSRLIK 540
541 EQTPAEFQSM LVNIFTHALE ALRRLYGHQK LLLDISLNEL ASVKSPNENQ HNMITQLDIR 600
601 TGINEIIRII QLRTGKIIAV RRELNLSLRY DYFLKFYAIC VIFIQECEVL SGEFLTKYLS 660
661 NVLASQIKHY ANAQSSKNYR NIKKKIDAEE WIPYIVDSSI QSDVNDIVSS IDIDPLSWTT 720
721 ILDMVGGSHD CENGRSEDKE KDEGNETYQG HRKSVVVGDK TFVASSSLLA TIEVIKELMV 780
781 LSINLPSIYL SNFEKLCYDA LQYYNSSAMA SVTQPGNSLL KTGRNLSIMG ESLDCLAEFV 840
841 IIVQRFYQRL SNSNRDFEPF DASHYTTLLG QFQASSNKIY MANAPPPPV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
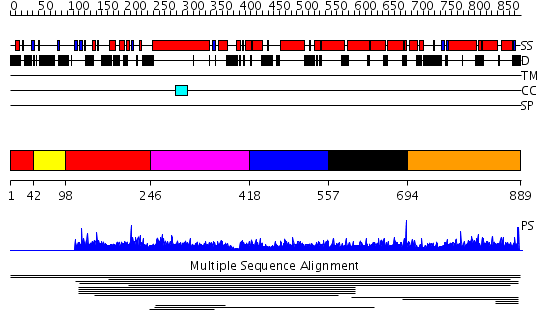
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..41] [98..245] |
7.08 | Botulinum neurotoxin | |
| 2 | View Details | [42..97] | 7.08 | Botulinum neurotoxin | |
| 3 | View Details | [246..417] | 8.47 | alpha-actinin | |
| 4 | View Details | [418..556] | 8.47 | alpha-actinin | |
| 5 | View Details | [557..693] | 8.47 | alpha-actinin | |
| 6 | View Details | [694..889] | 1.008939 | View MSA. Confident ab initio structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)