













| General Information: |
|
| Name(s) found: |
gi|220952014
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | synthetic construct |
| Length: | 921 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTDSKYFTTT KKGEIFELKS ELNNDKKEKK KEAVKKVIAS MTVGKDVSAL FPDVVNCMQT 60
61 DNLELKKLVY LYLMNYAKSQ PDMAIMAVNT FVKDCEDSNP LIRALAVRTM GCIRVDKITE 120
121 YLCEPLRKCL KDEDPYVRKT AAVCVAKLYD ISATMVEDQG FLDQLKDLLS DSNPMVVANA 180
181 VAALSEINEA SQSGQPLVEM NSVTINKLLT ALNECTEWGQ VFILDSLANY SPKDEREAQS 240
241 ICERITPRLA HANAAVVLSA VKVLMKLLEM LSSDSDFCAT LTKKLAPPLV TLLSSEPEVQ 300
301 YVALRNINLI VQKRPDILKH EMKVFFVKYN DPIYVKLEKL DIMIRLANQS NIAQVLSELK 360
361 EYATEVDVDF VRKAVRAIGR CAIKVEPSAE RCVSTLLDLI QTKVNYVVQE AIVVIKDIFR 420
421 KYPNKYESII STLCENLDTL DEPEARASMV WIIGEYAERI DNADELLDSF LEGFQDENAQ 480
481 VQLQLLTAVV KLFLKRPSDT QELVQHVLSL ATQDSDNPDL RDRGFIYWRL LSTDPAAAKE 540
541 VVLADKPLIS EETDLLEPTL LDELICHISS LASVYHKPPT AFVEGRGAGV RKSLPNRAAG 600
601 SAAGAEQAEN AAGSEAMVIP NQESLIGDLL SMDINAPAMP SAPAATSNVD LLGGGLDILL 660
661 GGPPAEAAPG GATSLLGDIF GLGGATLSVG VQIPKVTWLP AEKGKGLEIQ GTFSRRNGEV 720
721 FMDMTLTNKA MQPMTNFAIQ LNKNSFGLVP ASPMQAAPLP PNQSIEVSMA LGTNGPIQRM 780
781 EPLNNLQVAV KNNIDIFYFA CLVHGNVLFA EDGQLDKRVF LNTWKEIPAA NELQYTLSGV 840
841 IGTTDGIASK MTTNNIFTIA KRNVEGQDML YQSLKLTNNI WVLLELKLQP GNPEATLSLK 900
901 SRSVEVANII FAAYEAIIRS P |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
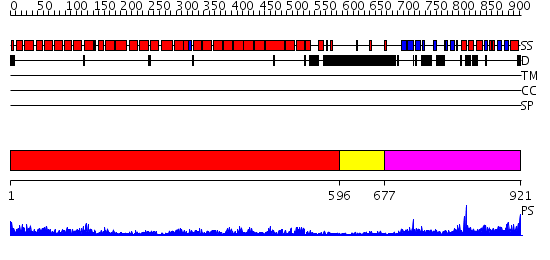
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..595] | 116.0 | AP1 clathrin adaptor core | |
| 2 | View Details | [596..676] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [677..921] | 55.30103 | Beta2-adaptin AP2 ear domain, N-terminal subdomain; Beta2-adaptin AP2, C-terminal subdomain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)