













| General Information: |
|
| Name(s) found: |
gi|207340890
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae AWRI1631 |
| Length: | 491 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRLISKRRIR FIVFILFGVL TVFVVSRLVV HFQYNQEIKF YKKYFQQRKD GLHEIYNPLE 60
61 IKQIPKETID DLYTARLDKE LKNGEVIEWS KFAYVNYVTN ADYLCNTLII FNDLKQEFET 120
121 KAKLVLLISK DLLDPNTSSN VAYISSLLNK IQAIDEDQVV IKLIDNIVKP KDTTPWNESL 180
181 TKLLVFNQTE FDRVIYLDND AILRSSLDEL FFLPNYIKFA APLTYWFLSN SDLEKSYHET 240
241 RHREKQPINL QSYTKVLTKR IGKGQMIYNH LPSLPHSLYL NSNNIAQDII SSTSSLSPLF 300
301 DFQSSKKVGK LKFASNLMVI NPSKEAFDEI VNVMLPKILN KKEKYDMDLI NEEMYNLKKI 360
361 IYKQFIFFRK VRKLFKPEVL VLPFARYGLL TGSLRNPRHY SIIYNDVLGY KTLDNDGNDI 420
421 PVGLNDSVAY SKYIHFSDYP LAKPWNYPSM KEFECIVKEE DAEDSKLEHQ ACDLWNSVYA 480
481 SYIQSREICL V |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
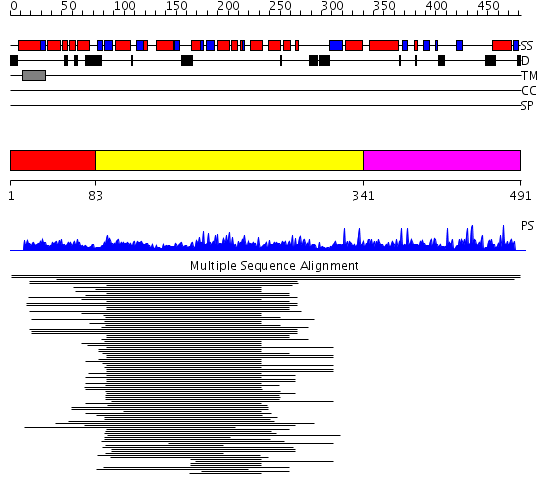
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [83..340] | 42.09691 | Glycogenin | |
| 3 | View Details | [341..491] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|