













| General Information: |
|
| Name(s) found: |
gi|41619520
[NCBI NR]
gi|18416582 [NCBI NR] gi|15375299 [NCBI NR] |
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 961 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
regulation of transcription
[IDA][TAS]
regulation of DNA endoreduplication [IMP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] transcription coactivator activity [IDA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEAESSTPQE RIPKLRHGRT SGPARRSTRG QWTAEEDEIL RKAVHSFKGK NWKKIAEYFK 60
61 DRTDVQCLHR WQKVLNPELV KGPWTKEEDE MIVQLIEKYG PKKWSTIARF LPGRIGKQCR 120
121 ERWHNHLNPA INKEAWTQEE ELLLIRAHQI YGNRWAELTK FLPGRSDNGI KNHWHSSVKK 180
181 KLDSYMSSGL LDQYQAMPLA PYERSSTLQS TFMQSNIDGN GCLNGQAENE IDSRQNSSMV 240
241 GCSLSARDFQ NGTINIGHDF HPCGNSQENE QTAYHSEQFY YPELEDISVS ISEVSYDMED 300
301 CSQFPDHNVS TSPSQDYQFD FQELSDISLE MRHNMSEIPM PYTKESKEST LGAPNSTLNI 360
361 DVATYTNSAN VLTPETECCR VLFPDQESEG HSVSRSLTQE PNEFNQVDRR DPILYSSASD 420
421 RQISEATKSP TQSSSSRFTA TAASGKGTLR PAPLIISPDK YSKKSSGLIC HPFEVEPKCT 480
481 TNGNGSFICI GDPSSSTCVD EGTNNSSEED QSYHVNDPKK LVPVNDFASL AEDRPHSLPK 540
541 HEPNMTNEQH HEDMGASSSL GFPSFDLPVF NCDLLQSKND PLHDYSPLGI RKLLMSTMTC 600
601 MSPLRLWESP TGKKTLVGAQ SILRKRTRDL LTPLSEKRSD KKLEIDIAAS LAKDFSRLDV 660
661 MFDETENRQS NFGNSTGVIH GDRENHFHIL NGDGEEWSGK PSSLFSHRMP EETMHIRKSL 720
721 EKVDQICMEA NVREKDDSEQ DVENVEFFSG ILSEHNTGKP VLSTPGQSVT KAEKAQVSTP 780
781 RNQLQRTLMA TSNKEHHSPS SVCLVINSPS RARNKEGHLV DNGTSNENFS IFCGTPFRRG 840
841 LESPSAWKSP FYINSLLPSP RFDTDLTIED MGYIFSPGER SYESIGVMTQ INEHTSAFAA 900
901 FADAMEVSIS PTNDDARQKK ELDKENNDPL LAERRVLDFN DCESPIKATE EVSSYLLKGC 960
961 R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
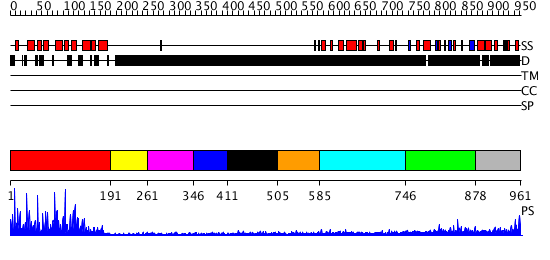
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 75.69897 | c-Myb, DNA-binding domain | |
| 2 | View Details | [191..260] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [261..345] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [346..410] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [411..504] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [505..584] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [585..745] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [746..877] | 2.281985 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [878..961] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)