













| General Information: |
|
| Name(s) found: |
gi|219556072
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Mycobacterium tuberculosis T17 |
| Length: | 1209 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVGAGWHFWV DRGGTFTDVV ARRPDGRLLT HKLLSDNPAR YRDAAVAGIR ALLANGEAGT 60
61 RVDAVRMGTT VATNALLERT GERTLLVITR GFGDALRIAY QNRPRIFDRR IVLPEMLYER 120
121 VVEVDERVTA DGRVLRAPDL EALGEKMRQA HADGIRAVAV VCLHSYLYPG HEREIGTLAQ 180
181 RIGFAQISLS SEVSPLMKLV PRGDTTVVDA YLSPVLRRYI NQVADQMRGV RLMFMQSNGG 240
241 LAQAGHFRGK DAILSGPAGG IVGMVRMSAL AGFDHVIGFD MGGTSTDVSH YAGEYERVFT 300
301 TQVAGVRLRA PMLDIHTVAA GGGSILHFDG SRYRVGPDSA GADPGPACYR GGGPLCVTDA 360
361 NVMLGRIQPT HFPSVFGPSG DQPLDAGTVR RGFTDLAADI AARTGDDRSP EQVAEGYLRI 420
421 AVANMANAVK KISVQKGHDV TRYALTTFGG AGGQHACAVA DALGIRTVLI PPMAGVLSAL 480
481 GIGLADTTAM REQSVEIPLG PAAPQRLASV AESLERAARA ELLDEGVPGE RIRVVRRVHL 540
541 RYEGTDTAIP VQLAEIETMA TAFESSHRAL YTFLLDRPLI AEAISVEATG LTDQPDLSQL 600
601 GDQANDTTGS SETVRIYSNG LWRDAPLRRR EAMRPGDVLT GPAIIAEANA TTVVDDGWQA 660
661 TMTETGHLLA QRVVTPPRPD AATRAGFEAG FEADPVLLEI FNNLFMSIAE QMGFRLEATA 720
721 QSVNIRERLD FSCALFDPDG NLVANAPHIP VHLGSMGTTV KEVIRRRLSG MKPGDVYAVN 780
781 DPYHGGTHLP DITVITPVFN TGGEDVLFFV ASRGHHAEIG GITPGSMPAD SREIHEEGVL 840
841 FDNWLLAENG RFREAETRRL LTEAPFGSRN PDTNLADLRA QIAANQKGVD EVGKMIDHFG 900
901 RDVVAAYMRH VQDNAEEAVR RVIDRLDNGA YRYRMDSGAT IAVRITVDRA ARSATIDFTG 960
961 TSAQLDTNFN APTSVVNAAV LYVFRTLVAD DIPLNDGCLR PLRIVVPEGS MLAPTHPAAV1020
1021 VAGNVETSQA ITGALFAALG VQAEGSGTMN NVTFGNERHQ YYETVGSGSG AGDGYHGASV1080
1081 VQTHMTNSRL TDPEVLEWRY PVLLREFAVR QGSGGAGRWR GGDGAVRRLE FTEPMTVSTL1140
1141 SGHRRVRPYG MAGGSPGELG RNRVERADGS TVELAGCGST HVEPGDTLVI ETPGGGGYGP1200
1201 ASTSARRRR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
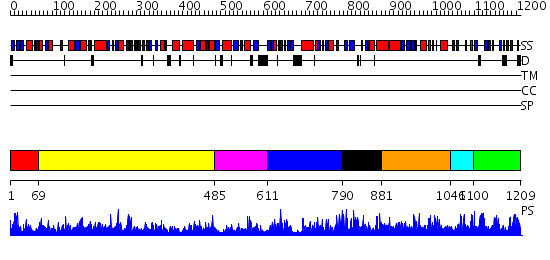
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..68] | 1.29 | Crystal Structure of Putative N-acetylglucosamine Kinase from Chromobacterium violaceum. Northeast Structural Genomics Target Cvr23. | |
| 2 | View Details | [69..484] | 46.69897 | Heat shock protein 70kDa, ATPase fragment | |
| 3 | View Details | [485..610] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [611..789] | 1.184954 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [790..880] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [881..1045] | 1.168955 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1046..1099] | 1.168991 | View MSA. No confident structure predictions are available. | |
| 8 | View Details | [1100..1209] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 5 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 6 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 7 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 8 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)