













| General Information: |
|
| Name(s) found: |
GACP4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 745 amino acids |
Gene Ontology: |
|
| Cellular Component: |
tubulin complex
[IPI]
|
| Biological Process: |
microtubule nucleation
[IMP]
cortical microtubule organization [IMP] growth [IMP] |
| Molecular Function: |
tubulin binding
[ISS]
protein binding [IPI] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLHELLLALL GFTGDLIVDE REQRKTLGLA FNSDSPLSDE CTFKLAPDIS FIEPSERDLI 60
61 ERLIKLGFYY RELDRFAKKS RNLSWIRSVT SVHPLERADE LSKQSREKKP SVYRRAIANG 120
121 IGEILSVYRS AVLHIEQKLL AETTPILATV TEGLNKFFVL FPPLYEVILE IERDDIRGGQ 180
181 LLNVLNKRCH CGVPELRTCL QRLLWNGHQV MYNQLAAWMV YGILQDPHGE FFIKRQDDGD 240
241 LDHRSSQEEV SEKLARTSVH ETSLTDWHSG FHISLDMLPD YIPMRLGESI LFAGKAIRVL 300
301 RNPSPAFQFQ KDKSFQQTMR GSQRIRGFMH SDFPETETEL DADLTGGELL PQSEADKIEA 360
361 MLKDLKESSE FHKRSFECTV DSVRAIAASH LWQLVVVRAD LNGHLKALKD YFLLEKGDFF 420
421 QCFLEESRQL MRLPPRQSTG ESDLMVPFQL AATKTIAEED KYFSRVSLRM PSFGVTVRSS 480
481 QADMVRSKVS LTGKANLTSD TSVDGWDAIA LEYSVDWPMQ LFFTQEVLSK YLKVFQYLIR 540
541 LKRTQMELEK SWASVMHQDH IESAQHRKDG LNGSTSQQRR QGIRPMWRVR EHMAFLIRNL 600
601 QFYIQVDVIE SQWKVLQTHI HDSQDFTELV GFHQEYLSAL ISQSFLDIGS VSRILDSIMK 660
661 LCLQFCWNIE NQESNPNTSE LENIAEEFNK KSNSLYTILR SSKLAGSQRA PFLRRFLLRL 720
721 NFNSFYEATA RGVLNVVRQR PALPL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
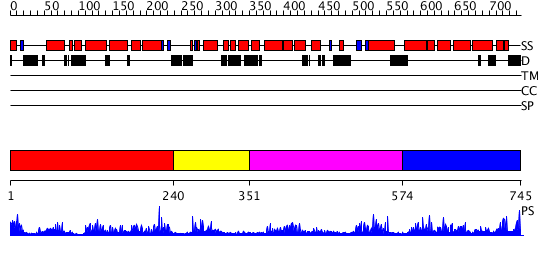
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..239] | 1.163981 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [240..350] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [351..573] | 2.197987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [574..745] | 2.18699 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.65 |
Source: Reynolds et al. (2008)