













| General Information: |
|
| Name(s) found: |
TOC90_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 793 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast outer membrane
[ISS]
intracellular [IEA] |
| Biological Process: | NONE FOUND |
| Molecular Function: |
GTP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKGFKDWVFA LSNSMASSRP LLGSDPFFRD PHQEQDNHSQ APAAPQPVTL SEPPCSTSSD 60
61 LEILPPLSQQ QVPLESLYQS SIDLNGKKHN PLAKIGGLQV QFLRLVQRFG QSQNNILVSK 120
121 VLYRVHLAML IRAEESELKN VKLRQDRAKA LAREQESSGI PELDFSLRIL VLGKTGVGKS 180
181 ATINSIFGQP KSETDAFRPG TDRIEEVMGT VSGVKVTFID TPGFHPLSSS STRKNRKILL 240
241 SIKRYVKKRP PDVVLYLDRL DMIDMRYSDF SLLQLITEIF GAAIWLNTIL VMTHSAATTE 300
301 GRNGQSVNYE SYVGQRMDVV QHYIHQAVSD TKLENPVLLV ENHPSCKKNL AGEYVLPNGV 360
361 VWKPQFMFLC VCTKVLGDVQ SLLRFRDSIG LGQPSSTRTA SLPHLLSVFL RRRLSSGADE 420
421 TEKEIDKLLN LDLEEEDEYD QLPTIRILGK SRFEKLSKSQ KKEYLDELDY RETLYLKKQL 480
481 KEECRRRRDE KLVEEENLED TEQRDQAAVP LPDMAGPDSF DSDFPAHRYR CVSAGDQWLV 540
541 RPVYDPQGWD RDVGFDGINI ETAAKINRNL FASATGQVSR DKQRFTIQSE TNAAYTRNFR 600
601 EQTFSVAVDL QSSGEDLVYS FQGGTKLQTF KHNTTDVGVG LTSFGGKYYV GGKLEDTLLV 660
661 GKRVKLTANA GQMRGSGQTA NGGSFEACIR GRDYPVRNEQ IGLTMTALSF KRELVLNYGL 720
721 QTQFRPARGT NIDVNINMNN RKMGKINVKL NSSEHWEIAL ISALTMFKAL VRRSKTEMTE 780
781 ENEEEKIVNF LVS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
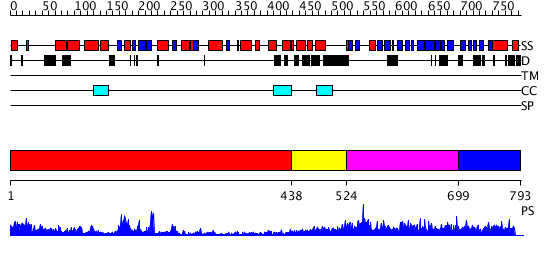
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..437] | 71.522879 | No description for 2hjgA was found. | |
| 2 | View Details | [438..523] | 28.154902 | No description for 2qagB was found. | |
| 3 | View Details | [524..698] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [699..793] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.48 |
Source: Reynolds et al. (2008)