













| General Information: |
|
| Name(s) found: |
PPR32_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 809 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
mRNA modification
[IMP]
|
| Molecular Function: |
endonuclease activity
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSQLVQFST VPQIPNPPSR HRHFLSERNY IPANVYEHPA ALLLERCSSL KELRQILPLV 60
61 FKNGLYQEHF FQTKLVSLFC RYGSVDEAAR VFEPIDSKLN VLYHTMLKGF AKVSDLDKAL 120
121 QFFVRMRYDD VEPVVYNFTY LLKVCGDEAE LRVGKEIHGL LVKSGFSLDL FAMTGLENMY 180
181 AKCRQVNEAR KVFDRMPERD LVSWNTIVAG YSQNGMARMA LEMVKSMCEE NLKPSFITIV 240
241 SVLPAVSALR LISVGKEIHG YAMRSGFDSL VNISTALVDM YAKCGSLETA RQLFDGMLER 300
301 NVVSWNSMID AYVQNENPKE AMLIFQKMLD EGVKPTDVSV MGALHACADL GDLERGRFIH 360
361 KLSVELGLDR NVSVVNSLIS MYCKCKEVDT AASMFGKLQS RTLVSWNAMI LGFAQNGRPI 420
421 DALNYFSQMR SRTVKPDTFT YVSVITAIAE LSITHHAKWI HGVVMRSCLD KNVFVTTALV 480
481 DMYAKCGAIM IARLIFDMMS ERHVTTWNAM IDGYGTHGFG KAALELFEEM QKGTIKPNGV 540
541 TFLSVISACS HSGLVEAGLK CFYMMKENYS IELSMDHYGA MVDLLGRAGR LNEAWDFIMQ 600
601 MPVKPAVNVY GAMLGACQIH KNVNFAEKAA ERLFELNPDD GGYHVLLANI YRAASMWEKV 660
661 GQVRVSMLRQ GLRKTPGCSM VEIKNEVHSF FSGSTAHPDS KKIYAFLEKL ICHIKEAGYV 720
721 PDTNLVLGVE NDVKEQLLST HSEKLAISFG LLNTTAGTTI HVRKNLRVCA DCHNATKYIS 780
781 LVTGREIVVR DMQRFHHFKN GACSCGDYW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
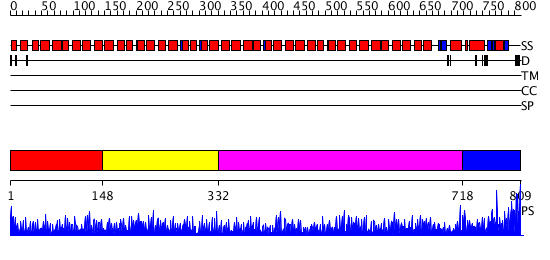
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..147] | 5.522879 | No description for 3cvqA was found. | |
| 2 | View Details | [148..331] | 10.69897 | No description for 2q7fA was found. | |
| 3 | View Details | [332..717] | 11.0 | No description for 2gw1A was found. | |
| 4 | View Details | [718..809] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)