













| General Information: |
|
| Name(s) found: |
DRL44_ARATH
[Swiss-Prot]
DRL10_ARATH [Swiss-Prot] |
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1017 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGELISFGI QNLWNLLSQE CELFQGVEDQ VTELKRDLNM LSSFLKDANA KKHTSAVVKN 60
61 CVEEIKEIIY DGEDTIETFV LEQNLGKTSG IKKSIRRLAC IIPDRRRYAL GIGGLSNRIS 120
121 KVIRDMQSFG VQQAIVDGGY KQPQGDKQRE MRQKFSKDDD SDFVGLEANV KKLVGYLVDE 180
181 ANVQVVSITG MGGLGKTTLA KQVFNHEDVK HQFDGLSWVC VSQDFTRMNV WQKILRDLKP 240
241 KEEEKKIMEM TQDTLQGELI RLLETSKSLI VLDDIWEKED WELIKPIFPP TKGWKVLLTS 300
301 RNESVAMRRN TSYINFKPEC LTTEDSWTLF QRIALPMKDA AEFKIDEEKE ELGKLMIKHC 360
361 GGLPLAIRVL GGMLAEKYTS HDWRRLSENI GSHLVGGRTN FNDDNNNTCN NVLSLSFEEL 420
421 PSYLKHCFLY LAHFPEDYEI KVENLSYYWA AEGIFQPRHY DGETIRDVGD VYIEELVRRN 480
481 MVISERDVKT SRFETCHLHD MMREVCLLKA KEENFLQITS SRPSTANLQS TVTSRRFVYQ 540
541 YPTTLHVEKD INNPKLRALV VVTLGSWNLA GSSFTRLELL RVLDLIEVKI KGGKLASCIG 600
601 KLIHLRYLSL EYAEVTHIPY SLGNLKLLIY LNLASFGRST FVPNVLMGMQ ELRYLALPSD 660
661 MGRKTKLELS NLVKLETLEN FSTENSSLED LCGMVRLSTL NIKLIEETSL ETLAASIGGL 720
721 KYLEKLEIYD HGSEMRTKEA GIVFDFVHLK RLWLKLYMPR LSTEQHFPSH LTTLYLESCR 780
781 LEEDPMPILE KLLQLKELEL GFESFSGKKM VCSSGGFPQL QRLSLLKLEE WEDWKVEESS 840
841 MPLLRTLDIQ VCRKLKQLPD EHLPSHLTSI SLFFCCLEKD PLPTLGRLVY LKELQLGFRT 900
901 FSGRIMVCSG GGFPQLQKLS IYRLEEWEEW IVEQGSMPFL HTLYIDDCPK LKKLPDGLQF 960
961 IYSLKNLKIS ERWKERLSEG GEEYYKVQHI PSVEFYHRVL HIFRSVGGDI TGRLLMR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
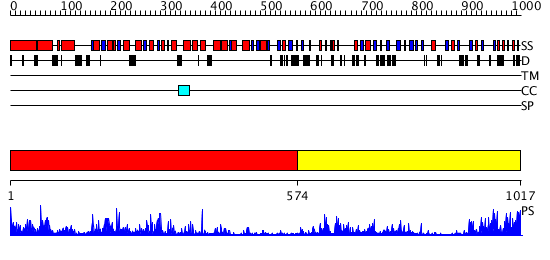
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..573] | 17.154902 | Structure of the apoptotic protease-activating factor 1 bound to ADP | |
| 2 | View Details | [574..1017] | 22.09691 | The molecular structure of toll-like receptor 3 ligand binding domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)