













| General Information: |
|
| Name(s) found: |
TAF1_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1919 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
DNA mediated transformation
[IMP]
|
| Molecular Function: |
DNA binding
[ISS]
histone acetyltransferase activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAESNGKGSH NETSSDDDDE YEDNSRGFNL GFIFGNVDNS GDLDADYLDE DAKEHLSALA 60
61 DKLGSSLPDI NLLAKSERTA SDPAEQDYDR KAEDAVDYED IDEEYDGPEV QVVSEEDHLL 120
121 PKKEYFSTAV ALGSLKSRAS VFDDEDYDEE EEQEEEQAPV EKSLETEKRE PVVLKEDKAL 180
181 EYEEEASILD KEDHMDTEDV QEEEVDELLE GTLDDKGATP LPTLYVEDGM VILQFSEIFA 240
241 IHEPPQKRDR RENRYVTCRD KYKSMDISEL VEDDEEVLLK SHGRIDTHVE QADLIQLDVP 300
301 FPIREGLQLV KASTIGGITP ESREFTKLGR DSCIMGELLK QDFIDDNSSL CQSQLSMQVF 360
361 PLDQHEWERR IIWEHSPEIS GNSGEIFEPG LEPEGMLVKG TNSETEQESL NVVNSRVQVQ 420
421 ADNNMFVPFS ANLLESFGSR GSQSTNESTN KSRHHPQLLR LESQWDENHL SGNDEAGVKK 480
481 IKRLEKDALG RFSRLVLRER DLGDEAWLDS IIWDSEKELS RSKLIFDLQD EQMVFEIFDN 540
541 EESKNLQLHA GAMIVSRSSK SKDETFQEGC ESNSGWQFNL SNDKFYMNGK SSQQLQANTN 600
601 KSSVHSLRVF HSVPAIKLQT MKSKLSNKDI ANFHRPKALW YPHDNELAIK QQGKLPTRGS 660
661 MKIIVKSLGG KGSKLHVGIE ESVSSLRAKA SRKLDFKETE AVKMFYKGKE LDDEKSLAAQ 720
721 NVQPNSLVHL IRTKVHLWPW AQKLPGENKS LRPPGAFKKK SDLSTKDGHV FLMEYCEERP 780
781 LMLSNAGMGA NLCTYYQKSS PEDQRGNLLR NQSDTLGNVM ILEPGDKSPF LGEIHAGCSQ 840
841 SSVETNMYKA PIFPQRLQST DYLLVRSPKG KLSLRRIDKI VVVGQQEPRM EVMSPGSKNL 900
901 QTYLVNRMLV YVYREFFKRG GGEHPIAADE LSFLFSNLTD AIIKKNMKII ACWKRDKNGQ 960
961 SYWTKKDSLL EPPESELKKL VAPEHVCSYE SMLAGLYRLK HLGITRFTLP ASISNALAQL1020
1021 PDEAIALAAA SHIERELQIT PWNLSSNFVA CTNQDRANIE RLEITGVGDP SGRGLGFSYV1080
1081 RAAPKAPAAA GHMKKKAAAG RGAPTVTGTD ADLRRLSMEA AREVLIKFNV PDEIIAKQTR1140
1141 WHRIAMIRKL SSEQAASGVK VDPTTIGKYA RGQRMSFLQM QQQAREKCQE IWDRQLLSLS1200
1201 AFDGDENESE NEANSDLDSF AGDLENLLDA EEGGEGEESN ISKNDKLDGV KGLKMRRRPS1260
1261 QVETDEEIED EATEYAELCR LLMQDEDQKK KKKKMKGVGE GMGSYPPPRP NIALQSGEPV1320
1321 RKANAMDKKP IAIQPDASFL VNESTIKDNR NVDSIIKTPK GKQVKENSNS LGQLKKVKIL1380
1381 NENLKVFKEK KSARENFVCG ACGQHGHMRT NKHCPRYREN TESQPEGIDM DKSAGKPSSS1440
1441 EPSGLPKLKP IKNSKAAPKS AMKTSVDEAL KGDKLSSKTG GLPLKFRYGI PAGDLSDKPV1500
1501 SEAPGSSEQA VVSDIDTGIK STSKISKLKI SSKAKPKESK GESERRSHSL MPTFSRERGE1560
1561 SESHKPSVSG QPLSSTERNQ AASSRHTISI PRPSLSMDTD QAESRRPHLV IRPPTEREQP1620
1621 QKKLVIKRSK EMNDHDMSSL EESPRFESRK TKRMAELAGF QRQQSFRLSE NSLERRPKED1680
1681 RVWWEEEEIS TGRHREVRAR RDYDDMSVSE EPNEIAEIRR YEEVIRSERE EEERQKAKKK1740
1741 KKKKKLQPEI VEGYLEDYPP RKNDRRLSER GRNVRSRYVS DFERDGAEYA PQPKRRKKGE1800
1801 VGLANILERI VDTLRLKEEV SRLFLKPVSK KEAPDYLDIV ENPMDLSTIR DKVRKIEYRN1860
1861 REQFRHDVWQ IKYNAHLYND GRNPGIPPLA DQLLEICDYL LDDYEDQLKE AEKGIDPND |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
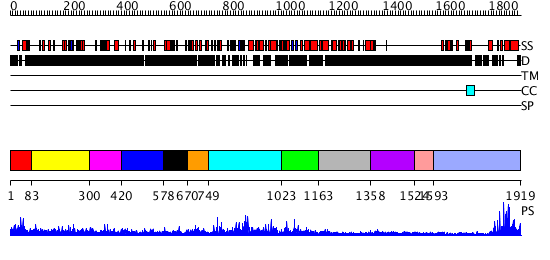
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..82] | 6.522879 | TAF(II)230 TBP-binding fragment | |
| 2 | View Details | [83..299] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [300..419] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [420..577] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [578..669] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [670..748] | 20.39794 | No description for 2ojrA was found. | |
| 7 | View Details | [749..1022] | 60.0 | No description for 2oanA was found. | |
| 8 | View Details | [1023..1162] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1163..1357] | N/A | No confident structure predictions are available. | |
| 10 | View Details | [1358..1523] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [1524..1592] | N/A | No confident structure predictions are available. | |
| 12 | View Details | [1593..1919] | 27.39794 | TAFII250 double bromodomain module |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)