













| General Information: |
|
| Name(s) found: |
SCL14_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 769 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA]
cytosol [IDA] |
| Biological Process: |
positive regulation of gene-specific transcription
[IMP]
response to xenobiotic stimulus [IMP] regulation of transcription [TAS] |
| Molecular Function: |
transcription factor activity
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGSYPDGFPG SMDELDFNKD FDLPPSSNQT LGLANGFYLD DLDFSSLDPP EAYPSQNNNN 60
61 NNINNKAVAG DLLSSSSDDA DFSDSVLKYI SQVLMEEDME EKPCMFHDAL ALQAAEKSLY 120
121 EALGEKYPSS SSASSVDHPE RLASDSPDGS CSGGAFSDYA STTTTTSSDS HWSVDGLENR 180
181 PSWLHTPMPS NFVFQSTSRS NSVTGGGGGG NSAVYGSGFG DDLVSNMFKD DELAMQFKKG 240
241 VEEASKFLPK SSQLFIDVDS YIPMNSGSKE NGSEVFVKTE KKDETEHHHH HSYAPPPNRL 300
301 TGKKSHWRDE DEDFVEERSN KQSAVYVEES ELSEMFDKIL VCGPGKPVCI LNQNFPTESA 360
361 KVVTAQSNGA KIRGKKSTST SHSNDSKKET ADLRTLLVLC AQAVSVDDRR TANEMLRQIR 420
421 EHSSPLGNGS ERLAHYFANS LEARLAGTGT QIYTALSSKK TSAADMLKAY QTYMSVCPFK 480
481 KAAIIFANHS MMRFTANANT IHIIDFGISY GFQWPALIHR LSLSRPGGSP KLRITGIELP 540
541 QRGFRPAEGV QETGHRLARY CQRHNVPFEY NAIAQKWETI QVEDLKLRQG EYVVVNSLFR 600
601 FRNLLDETVL VNSPRDAVLK LIRKINPNVF IPAILSGNYN APFFVTRFRE ALFHYSAVFD 660
661 MCDSKLARED EMRLMYEKEF YGREIVNVVA CEGTERVERP ETYKQWQARL IRAGFRQLPL 720
721 EKELMQNLKL KIENGYDKNF DVDQNGNWLL QGWKGRIVYA SSLWVPSSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
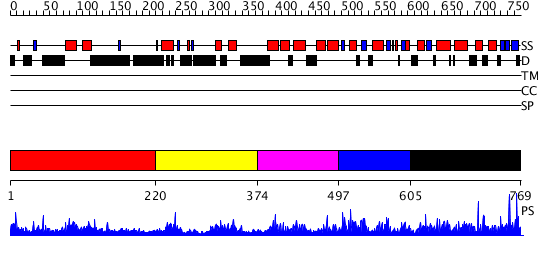
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..219] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [220..373] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [374..496] | 5.116988 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [497..604] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [605..769] | 4.124959 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)