













| General Information: |
|
| Name(s) found: |
LHY_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 11 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 645 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
response to gibberellin stimulus
[IEP]
response to abscisic acid stimulus [IEP] response to salt stress [IEP] long-day photoperiodism, flowering [IGI] response to jasmonic acid stimulus [IEP] response to cadmium ion [IEP] response to salicylic acid stimulus [IEP] regulation of transcription [TAS] response to ethylene stimulus [IEP] response to auxin stimulus [IEP] |
| Molecular Function: |
DNA binding
[ISS]
transcription factor activity [ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTNTSGEEL LAKARKPYTI TKQRERWTED EHERFLEALR LYGRAWQRIE EHIGTKTAVQ 60
61 IRSHAQKFFT KLEKEAEVKG IPVCQALDIE IPPPRPKRKP NTPYPRKPGN NGTSSSQVSS 120
121 AKDAKLVSSA SSSQLNQAFL DLEKMPFSEK TSTGKENQDE NCSGVSTVNK YPLPTKQVSG 180
181 DIETSKTSTV DNAVQDVPKK NKDKDGNDGT TVHSMQNYPW HFHADIVNGN IAKCPQNHPS 240
241 GMVSQDFMFH PMREETHGHA NLQATTASAT TTASHQAFPA CHSQDDYRSF LQISSTFSNL 300
301 IMSTLLQNPA AHAAATFAAS VWPYASVGNS GDSSTPMSSS PPSITAIAAA TVAAATAWWA 360
361 SHGLLPVCAP APITCVPFST VAVPTPAMTE MDTVENTQPF EKQNTALQDQ NLASKSPASS 420
421 SDDSDETGVT KLNADSKTND DKIEEVVVTA AVHDSNTAQK KNLVDRSSCG SNTPSGSDAE 480
481 TDALDKMEKD KEDVKETDEN QPDVIELNNR KIKMRDNNSN NNATTDSWKE VSEEGRIAFQ 540
541 ALFARERLPQ SFSPPQVAEN VNRKQSDTSM PLAPNFKSQD SCAADQEGVV MIGVGTCKSL 600
601 KTRQTGFKPY KRCSMEVKES QVGNINNQSD EKVCKRLRLE GEAST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
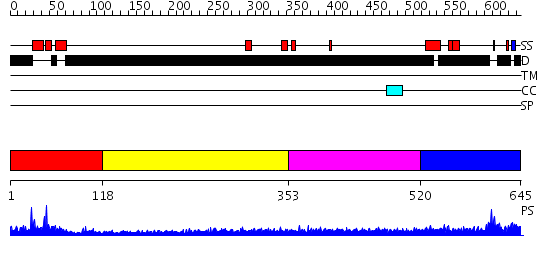
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..117] | 11.69897 | No description for 2v1dB was found. | |
| 2 | View Details | [118..352] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [353..519] | 3.185995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [520..645] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.45 |
Source: Reynolds et al. (2008)