













| General Information: |
|
| Name(s) found: |
gi|22328601
[NCBI NR]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 263 amino acids |
Gene Ontology: |
|
| Cellular Component: |
intracellular
[IEA]
|
| Biological Process: |
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
[IEA]
|
| Molecular Function: |
nucleic acid binding
[IEA]
3'-5' exonuclease activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTTEELKIS HYKLYKCFDF LVVANNKIEV AQAFVKYRRP EYFLPLVGVE FVSLTLVGKH 60
61 FKLGKYSVVV LRGEVHSETL RCLLPKLTDQ PIGSSFLLLR PFRDVGNYCL LLHILPSSKK 120
121 FTLAMQFLCE LIDSTSKEEE DVFRVLVYAS DELLDVKTYR ALIGCSWELF RCSPGEGCNC 180
181 PDMCRLQHLI EDSTLVKVGI GIDGDSVKLF HDYGLLKPNR IRLGNWEVHP LSKQQLQYAA 240
241 TDAYASWHLH QVLKDLPDAV SGS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
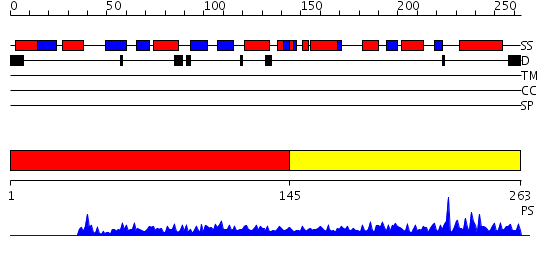
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..144] | 1.113971 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [145..263] | 24.39794 | Crystal Structure of Escherichia coli RNase D, an exoribonuclease involved in structured RNA processing |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)