













| General Information: |
|
| Name(s) found: |
EDR2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 718 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSKVVYEGWM VRYGRRKIGR SYIHMRYFVL EPRLLAYYKK KPQDYQVPIK TMLIDGNCRV 60
61 EDRGLKTHHG HMVYVLSVYN KKEKSHRITM AAFNIQEALM WKEKIESVID QHQESQVPNG 120
121 QQYVSFEYKS GMDTGRTASS SDHESQFSAA EDEEDSRRSL MRRTTIGNGP PESVLDWTKE 180
181 FDAELANQNS DNQAFSRKHW RLLQCQNGLR IFEELLEVDY LPRSCSRAMK AVGVVEATCE 240
241 EIFELLMSMD GTRYEWDCSF QFGSLVEEVD GHTAVLYHRL LLDWFPMIVW PRDLCYVRYW 300
301 RRNDDGSYVV LFRSREHENC GPQPGCVRAH LESGGYNISP LKPRNGRPRT QVQHLIQIDL 360
361 KGWGAGYLPA FQQHCLLQML NSVAGLREWF SQTDERGVHT RIPVMVNMAS SSLSLTKSGK 420
421 SLHKSAFSLD QTNSVNRNSL LMDEDSDDDD EFQIAESEQE PETSKPETDV KRPEEEPAHN 480
481 IDLSCFSGNL KRNENENARN CWRISDGNNF KVRGKNFGQE KRKIPAGKHL MDLVAVDWFK 540
541 DSKRIDHVAR RKGCAAQVAA EKGLFSMVVN VQVPGSTHYS MVFYFVMKEL VPGSLLQRFV 600
601 DGDDEFRNSR LKLIPLVPKG SWIVRQSVGS TPCLLGKAVD CNYIRGPTYL EIDVDIGSST 660
661 VANGVLGLVI GVITSLVVEM AFLVQANTAE EQPERLIGAV RVSHIELSSA IVPNLESE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
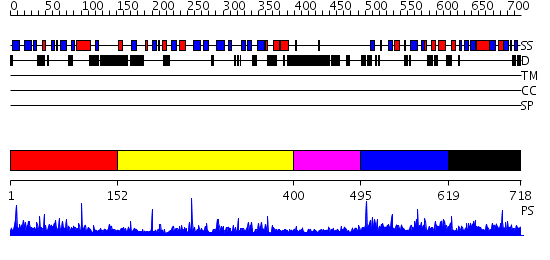
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..151] | 5.28 | No description for 2ys3A was found. | |
| 2 | View Details | [152..399] | 33.39794 | Cholesterol-regulated Start protein 4 (Stard4). | |
| 3 | View Details | [400..494] | 2.144992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [495..618] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [619..718] | 2.02997 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.73 |
Source: Reynolds et al. (2008)