













| General Information: |
|
| Name(s) found: |
gi|79315061
[NCBI NR]
gi|30694123 [NCBI NR] gi|222424068 [NCBI NR] |
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 733 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
signal transduction
[ISS]
|
| Molecular Function: |
phosphoinositide binding
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGVSQTDGRM EGWLYTIRHN RFGLQFSRKR YFVLHENNLT SFKSVPSDHN EEPERRASLD 60
61 CCIRVTDNGR ESFHRKILFI FTLYNTSNHL DQLKLGASSP EEAAKWIRSL QDASQKGFPI 120
121 PDCEFFVSHA EKGLVKLDVS KRNRRKNSVD WTNYSSTNYS STSLNVETNV APDVIAPSPW 180
181 KIFGCQNGLR LFKEAKDWDS RGRHWDDHPA IMAVGVIDGT SEDIFNTLMS LGPLRSEWDF 240
241 CFYKGNVVEH LDGHTDIIHL QLYSDWLPWG MNRRDLLLRR YWRREDDGTY VILCHSVYHK 300
301 NCPPKKGYVR ACVKSGGYVV TPANNGKQSL VKHMVAIDWR SWNLYMRPSS ARSITIRVVE 360
361 RVAALREMFK AKQGHGFTEF VSGEFLDTKP CLSKINTMPL KTEAKEVDLE TMHAEEMDKP 420
421 TSARNSLMDL NDASDEFFDV PEPNESTEFD SFIDSSPYSQ GHQLKIPTPA GIVKKLQDLA 480
481 INKKGYMDLQ EVGLEENNTF FYGATLQKDP SLTLPCSWST ADPSTFLIRG NNYLKNQQKV 540
541 KAKGTLMQMI GADWISSDKR EDDLGGRIGG LVQEYAAKGS PEFFFIVNIQ VPGSAMYSLA 600
601 LYYMLKTPLE EHPLLESFVN GDDAYRNSRF KLIPHISKGS WIVKQSVGKK ACLVGQVLEV 660
661 CYTRGKNYLE LDIDVGSSTV ARGVTNLVLG YLNNLVIEMA FLIQANTVEE LPELLLGTCR 720
721 LNYLDVSKSV KER |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
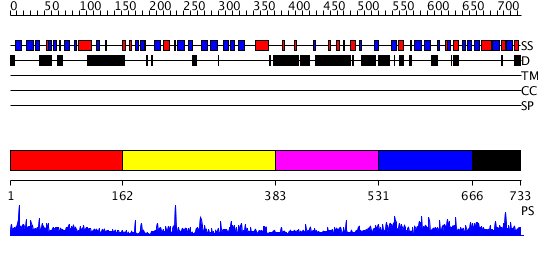
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..161] | 5.07 | Crystal Structure of the PH Domain of SKAP-Hom with 8 Vector-derived N-terminal Residues | |
| 2 | View Details | [162..382] | 34.0 | No description for 2psoA was found. | |
| 3 | View Details | [383..530] | 2.143981 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [531..665] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [666..733] | 4.030995 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)