













| General Information: |
|
| Name(s) found: |
SPP3A_ARATH
[Swiss-Prot]
|
| Description(s) found:
SHOW ONLY BEST |
|
| Organism: | Arabidopsis thaliana |
| Length: | 425 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
sucrose biosynthetic process
[IEA][ISS]
metabolic process [IEA] |
| Molecular Function: |
magnesium ion binding
[IEA]
catalytic activity [IEA] phosphatase activity [IEA] sucrose-phosphatase activity [IEA][ISS] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDRLEGPPRL ILVADLDCTL VDHDDPENND LLRFNALWEA HYRHDSLLVY CTGRSFSSYS 60
61 SLRKKRPLLT PDIAVTSVGS EIVYGGGEST VSDVVWTARL DYKWNRDIVV EETLKFPKLE 120
121 PQPDKSQEEH KVSFFVGRED AVEIMKVLPG ILEERGVDVK LVYSNGYAFD VLPRGAGKQG 180
181 ALTYLLDKLD IEGKQPSNTL VCGDSGNDAE LFNISDVYGV MVSNSHEELL QWYEENAKDN 240
241 PKIFHASERC GAGMIEAIQR FNLGPNVSPR DVMDTENFHG ESLNPAHEVV QFYLFYERWR 300
301 CGEVEKSDKY LQNLKSLSSP LGIFVHPSGV EKPIHEWIDE MENLYGDGKE KKFRIWLDNV 360
361 TSSHISSDTW LAKFVKHELS EGKVRSCSTK VLLSYKEEKQ RLTWMHIHQS WLDESSSDDQ 420
421 EKWIF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
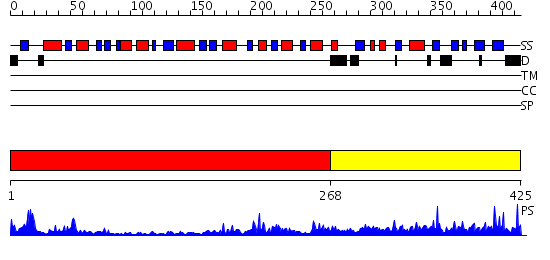
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..267] | 34.221849 | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | |
| 2 | View Details | [268..425] | 5.22 | Structural Genomics, 1.5A crystal structure of a hypothetical protein PA1314 from Pseudomonas aeruginosa |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 |
|
|||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)