













| General Information: |
|
| Name(s) found: |
gi|30683868
[NCBI NR]
gi|30683865 [NCBI NR] gi|222422950 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 918 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPLRLDIKRK FAQRSERVKS VDLHPTEPWI LASLYSGTVC IWNYQTQTIT KSFEVTELPV 60
61 RSAKFIPRKQ WVVAGADDMY IRVYNYNTMD KVKVFEAHSD YIRCVAVHPT LPYVLSSSDD 120
121 MLIKLWDWEN GWACTQIFEG HSHYVMQVVF NPKDTNTFAS ASLDRTIKIW NLGSPDPNFT 180
181 LDAHQKGVNC VDYFTGGDKP YLITGSDDHT AKVWDYQTKS CVQTLDGHTH NVSAVCFHPE 240
241 LPIIITGSED GTVRIWHATT YRLENTLNYG LERVWAIGYI KSSRRVVIGY DEGTIMVKLG 300
301 REIPVASMDS SGKIIWAKHN EIQTANIKSI GAGYEATDGE RLPLSVKELG TCDLYPQSLK 360
361 HNPNGRFVVV CGDGEYIIYT ALAWRNRSFG SGLEFVWSSE GECAVRESSS KIKIFSKNFQ 420
421 ERKSIRPTFS AEKIFGGTLL AMCSNDFICF YDWAECRLIQ QIDVTVKNLY WAESGDLVAI 480
481 ASDTSFYILK YNRELVSSHF DSGRPTDEEG VEDAFEVLHE NDERVRTGIW VGDCFIYNNS 540
541 SWKLNYCVGG EVTTMYHLDR PMYLLGYIAN QSRVYLVDKE FNVIGYTLLL SLIEYKTLVM 600
601 RGDLDRANQI LPTIPKEQHN NVAHFLESRG MIEDALEIAT DPDYKFDLAI QLGRLEIAKE 660
661 IAEEVQSESK WKQLGELAMS SGKLQLAEDC MKYAMDLSGL LLLYSSLGDA EGVSKLACLA 720
721 KEQGKNNVAF LCLFTLGRLE DCLQLLVESN RIPEAALMAR SYLPSKVSEI VALWREDLSK 780
781 VNPKAAESLA DPEEYSNLFE DWQVALSVEA NTAETRGVYT AAENYPSHAD KPSITLVEAF 840
841 RNLQVEAEES LENGNIDHEV AEENGHVENE GDEEEQQEEE VNEEEGVVDA DSTDGAVLVN 900
901 GSEGEEEWGT NNKGNPSA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
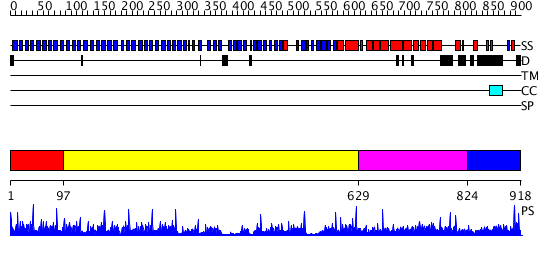
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..96] | 83.69897 | PAF-AH Holoenzyme: Lis1/Alfa2 | |
| 2 | View Details | [97..628] | 58.154902 | Actin interacting protein 1 | |
| 3 | View Details | [629..823] | 1.42 | Clathrin heavy chain proximal leg segment | |
| 4 | View Details | [824..918] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.88 |
Source: Reynolds et al. (2008)