













| General Information: |
|
| Name(s) found: |
gi|30678104
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 871 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIMNGKLKSR YNYLLKAGYG SSSDVIWRPG PQFSLSVPSS VNQDRKIIIR DSWMSMSISS 60
61 KSQESYGWGS WIDDAYLFPN CVTPAQSEDE CTSADSAIEV PRTHLNDKQV GAESFLCDEL 120
121 AAFSSENSNL SALFSDNYQP IEEPWLIQES ITLQHERNMQ TDSEQDVESC DDNENNLNTD 180
181 EQNHQLTETL LPDGGFFQSE SIATTILINS SICTVQRIAV LEGGKLVELL LEPVKTNVQC 240
241 DSVYLGVITK FVPHMGGAFV NIGSARHSFM DIKSNREPFI FPPFCDGSKK QAADGSPILS 300
301 MNDIPAPHEI EHASYDFEAS SLLDIDSNDP GESFHDDDDE HENDEYHVSD HLAGLVNGTV 360
361 VNHGAVEVGS ENGHIPMERG HSADSLDSNA SVAKASKVMS SKDNKWIQVR KGTKIIVQVV 420
421 KEGLGTKGPT LTAYPKLRSR FWVLLTRCKR IGVSKKISGV ERTRLKVIAK TLQPQGFGLT 480
481 VRTVAAGHSL EELQKDLDGL LLTWKNITDE AKSAALAADE GVEGAIPALL HRAMGQTLSV 540
541 VQDYFNDKVE KMVVDSPRTY HEVTHYLQDM APDLCNRVEL HDKGIPLFDL YEIEEEIEGI 600
601 LSKRVPLSNG GSLVIEQTEA LVSIDVNGGH GMFGQGNSQE KAILEVNLAA ARQIAREIRL 660
661 RDIGGIIVVD FIDMADESNK RLVYEEVKKA VERDRSLVKV SELSRHGLME ITRKRVRPSV 720
721 TFMISEPCSC CHATGRVEAL ETTFSKIEQE ICRQLAKMEK RGDLENPKSW PRFILRVDSH 780
781 MSSFLTTGKR TRLAILSSSL KVWILLKVAR HFTRGTFEVK PFMDEKTVNE RQHQVAISLL 840
841 KKADAIADSS GKKKLTLIPI KKEKTSGKQR R |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
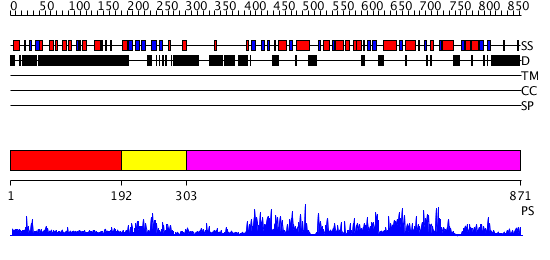
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..191] | 26.062997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [192..302] | 2.075987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [303..871] | 163.0 | Catalytic domain of E. coli RNase E |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)