













| General Information: |
|
| Name(s) found: |
gi|42569389
[NCBI NR]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 1106 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRTTFLEDA ERVKLYNDAR ALFANGDHIK ALEIIEDIIL VHKEDEKSWI LHSEQGEMFN 60
61 DLAGKAENPY VDFTYMLGSV GCYSKDVLLS RPWAHGLHYL AQESGSVMYY KKCVNVAKQG 120
121 LSVTFPDASE SFAQLLLENL KNDMKENLET LIKDAESNIA DSKTVELKGL LQQDFEPEVL 180
181 ESKKRPELVK NEFKGLRSFW LGLDVNVKRD FMKVSIAKLI GFVKGVHHRP GRDALKQILD 240
241 SAKEDRKWTF WMCRTKCSKK FSSAEECKNH LEKEHAADYK PFSEMDMAKR IGKDWTRKIS 300
301 LGSWEPVDIA AAVEMIKNRL PEVKAFSYKN GWSKEWPLAE DEERSKLLKE IKFFLVMFCD 360
361 LKILPCSIRD WMKRYSVTHL KNLNVSEQSL VDSHLVDTPQ SICFLECHKL SYILNLLKRI 420
421 KCERDDGTNL VRRAVDSILD STRSKEKIDF DPQFSFLLLD RRLLKSNNGP FVDEGEINVF 480
481 DPNVHYAKAH AQGDDIVSWL TDYNSVEKTF PRPIREHNLD IWVAVLRAVQ FTCRTFGTKY 540
541 AKKVQLFDYL AAIVAAEKLC KSEYERRRKL KEDQWNSYAS LLCDKCEECF SRKNFLKRKL 600
601 FLSVVRDILE EASDLTFDLP DIEGCMNLIR EHKSLNDDIV LKSLICLNSV VIFKILLIDS 660
661 KILLVDNSRI SLLNNLTRLS LFDNRTYILQ LLKPFLLNEI VNMESKAKSD AAVADLLLEE 720
721 EKKNPKPSPI QSKKKKNTSK KRNSTSMSSP LSKPGEHLEP ETTSPTVEED SMELGDTVNQ 780
781 EEVMKNMLGE DSQSEHLESA LSEASARYNS ALDMTLKALL NIKILKEDLM HNMQPLQHHL 840
841 EDQVPSVLQN FFTAFVSEEI KTEGVYSYLL SDLLALLEEV LSMSSGAAEV LVAILEFWHC 900
901 WKNAERESLV TRLFTLEEKE RMSCRKCRKK PNYPEQSSYG IVMAADSIRD LKCALGNIEF 960
961 VDILKVIRMD YTMLCDIKNG GCGIANFIHH IISKCPPIFT IVLEWEKSET EKEISETTKA1020
1021 FEWEIDISRL YAGLEPKTNY RLASMVGCDE KQEHICIAYE KNRWVNLRRD ALAGEDVGNW1080
1081 KSVVRFCGER KVRPEILFYE VVRSMA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
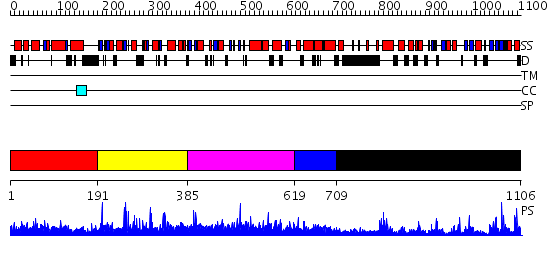
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] | 2.15 | TETRATRICOPEPTIDE REPEATS OF PROTEIN PHOSPHATASE 5 | |
| 2 | View Details | [191..384] | 1.020968 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [385..618] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [619..708] | N/A | Confident ab initio structure predictions are available. | |
| 5 | View Details | [709..1106] | 52.09691 | Crystal structure of HAUSP |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)