GO TERM SUMMARY
|
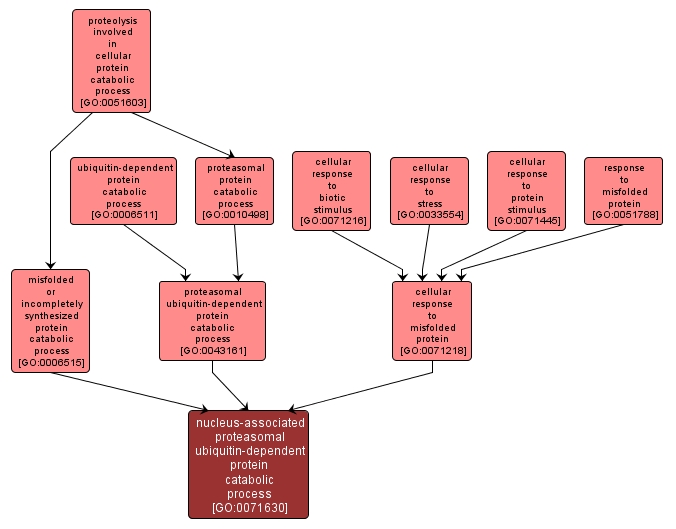
| Name: |
nucleus-associated proteasomal ubiquitin-dependent protein catabolic process |
| Acc: |
GO:0071630 |
| Aspect: |
Biological Process |
| Desc: |
The chemical reactions and pathways resulting in the breakdown of misfolded proteins via a mechanism in which the proteins are transported to the nucleus for ubiquitination, and then targeted to proteasomes for degradation. |
Synonyms:
- nucleus-associated proteasomal ubiquitin-dependent protein breakdown
- nucleus-associated proteasomal ubiquitin-dependent protein catabolism
- nucleus-associated proteasomal ubiquitin-dependent protein degradation
|