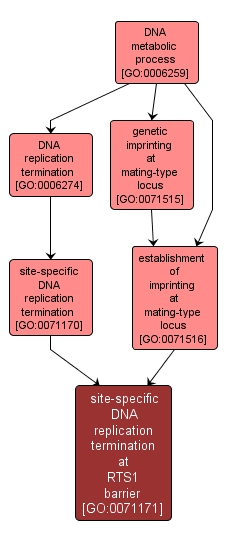
GO TERM SUMMARY
|
| Name: |
site-specific DNA replication termination at RTS1 barrier |
| Acc: |
GO:0071171 |
| Aspect: |
Biological Process |
| Desc: |
A DNA replication termination process that takes place at the RTS1 termination site in the mating type locus, in a specific direction required for subsequent imprinting and mating-type switching. |
|

|
INTERACTIVE GO GRAPH
|