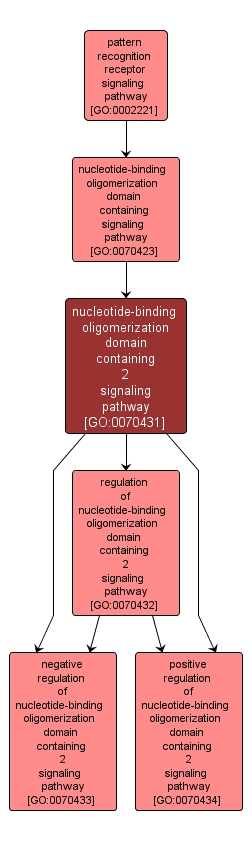
GO TERM SUMMARY
|
| Name: |
nucleotide-binding oligomerization domain containing 2 signaling pathway |
| Acc: |
GO:0070431 |
| Aspect: |
Biological Process |
| Desc: |
Any series of molecular signals generated as a consequence of binding to nucleotide-binding oligomerization domain containing 2 (NOD2). |
Synonyms:
- nucleotide-binding oligomerization domain containing 2 signalling pathway
- NOD2 signaling pathway
|
|

|
INTERACTIVE GO GRAPH
|