GO TERM SUMMARY
|
| Name: |
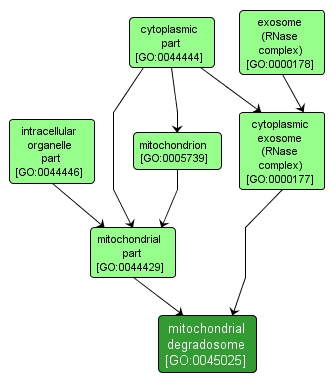
mitochondrial degradosome |
| Acc: |
GO:0045025 |
| Aspect: |
Cellular Component |
| Desc: |
The mitochondrial degradosome (mtEXO) is a three-protein complex which has a 3' to 5' exoribonuclease activity and participates in intron-independent turnover and processing of mitochondrial transcripts. |
|

|
INTERACTIVE GO GRAPH
|