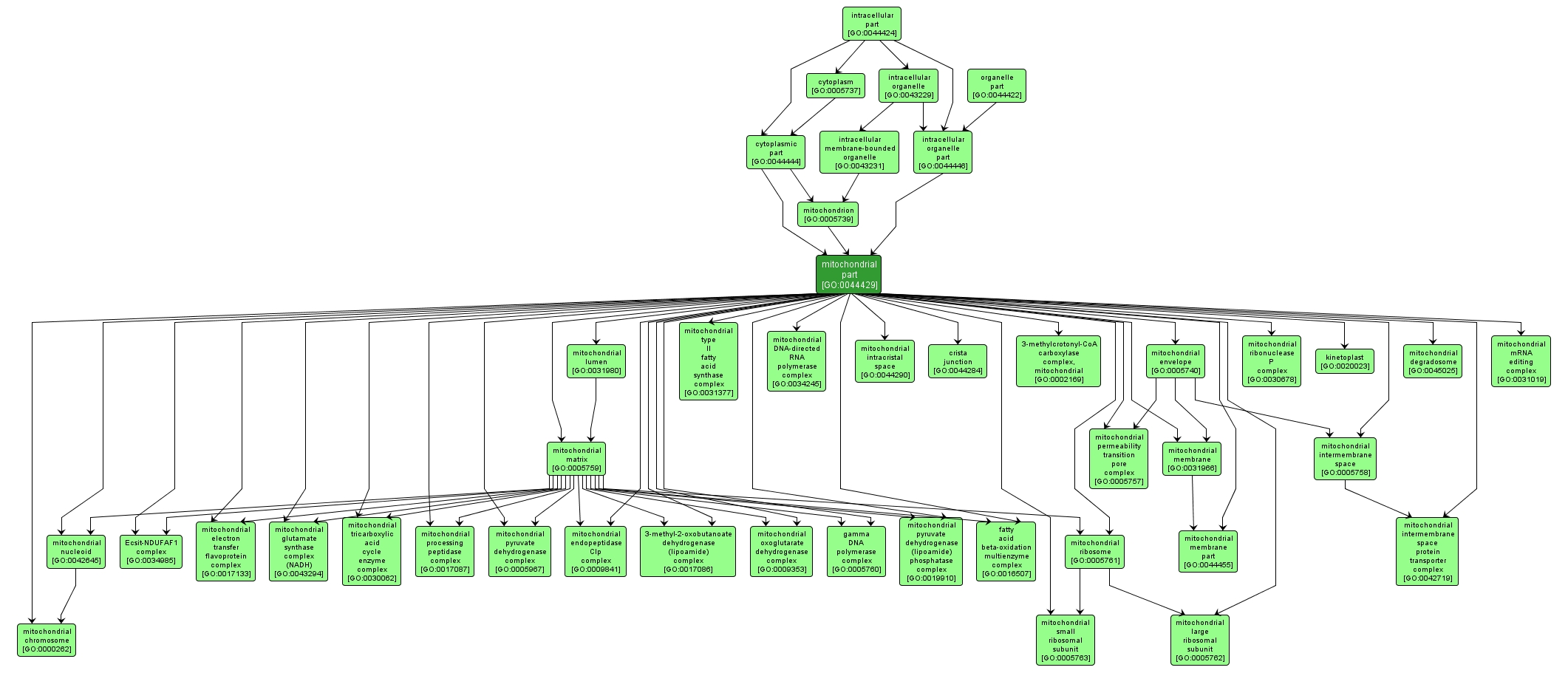
GO TERM SUMMARY
|
| Name: |
mitochondrial part |
| Acc: |
GO:0044429 |
| Aspect: |
Cellular Component |
| Desc: |
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|