GO TERM SUMMARY
|
| Name: |
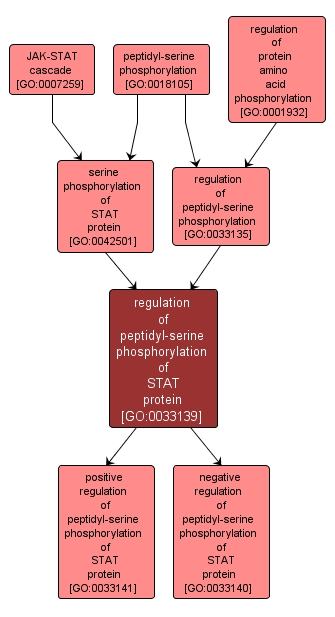
regulation of peptidyl-serine phosphorylation of STAT protein |
| Acc: |
GO:0033139 |
| Aspect: |
Biological Process |
| Desc: |
Any process that modulates the frequency, rate or extent of the phosphorylation of a serine residue of a STAT (Signal Transducer and Activator of Transcription) protein. |
|

|
INTERACTIVE GO GRAPH
|