GO TERM SUMMARY
|
| Name: |
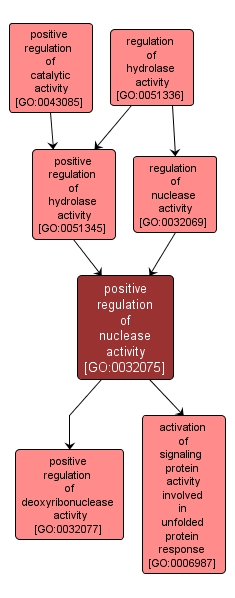
positive regulation of nuclease activity |
| Acc: |
GO:0032075 |
| Aspect: |
Biological Process |
| Desc: |
Any process that activates or increases the frequency, rate or extent of nuclease activity, the hydrolysis of ester linkages within nucleic acids. |
Synonyms:
- upregulation of nuclease activity
- stimulation of nuclease activity
- up-regulation of nuclease activity
- nuclease activator
- up regulation of nuclease activity
- activation of nuclease activity
|
|

|
INTERACTIVE GO GRAPH
|