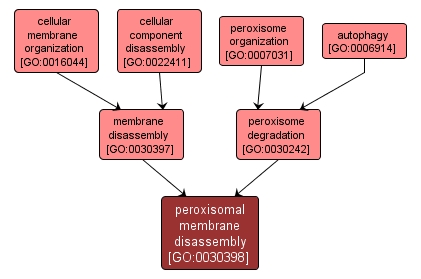
GO TERM SUMMARY
|
| Name: |
peroxisomal membrane disassembly |
| Acc: |
GO:0030398 |
| Aspect: |
Biological Process |
| Desc: |
The controlled breakdown of the membranes of cargo-carrying vesicles formed during peroxisome degradation. |
Synonyms:
- peroxisomal membrane degradation
- peroxisomal membrane breakdown
- peroxisomal membrane catabolism
|
|

|
INTERACTIVE GO GRAPH
|