GO TERM SUMMARY
|
| Name: |
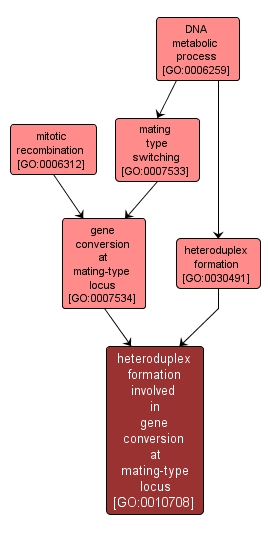
heteroduplex formation involved in gene conversion at mating-type locus |
| Acc: |
GO:0010708 |
| Aspect: |
Biological Process |
| Desc: |
The formation of a stable duplex DNA that contains one strand from each of the two recombining DNA molecules resulting in the conversion of the mating-type locus from one allele to another. |
|

|
INTERACTIVE GO GRAPH
|