GO TERM SUMMARY
|
| Name: |
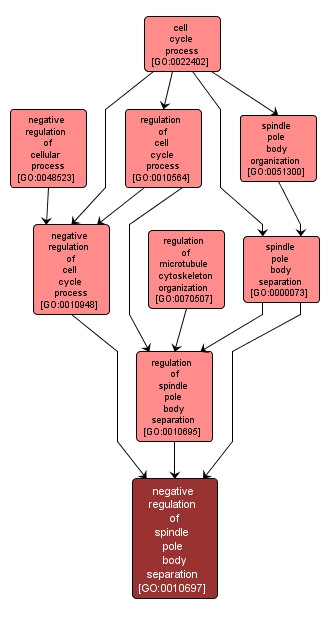
negative regulation of spindle pole body separation |
| Acc: |
GO:0010697 |
| Aspect: |
Biological Process |
| Desc: |
Any process that decreases the rate, frequency or extent of the process involving the release of duplicated spindle pole bodies (SPBs) and their migration away from each other within the nuclear membrane. |
Synonyms:
- negative regulation of SPB separation
|
|

|
INTERACTIVE GO GRAPH
|