GO TERM SUMMARY
|
| Name: |
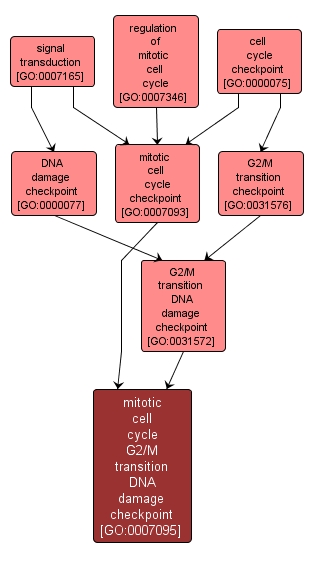
mitotic cell cycle G2/M transition DNA damage checkpoint |
| Acc: |
GO:0007095 |
| Aspect: |
Biological Process |
| Desc: |
A signal transduction-based surveillance mechanism that acts during a mitotic cell cycle to ensure accurate chromosome segregation by preventing entry into mitosis in the presence of damaged DNA. |
| Synonyms:
|
|

|
INTERACTIVE GO GRAPH
|