GO TERM SUMMARY
|
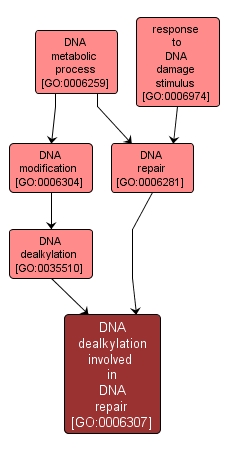
| Name: |
DNA dealkylation involved in DNA repair |
| Acc: |
GO:0006307 |
| Aspect: |
Biological Process |
| Desc: |
The repair of alkylation damage, e.g. the removal of the alkyl group at the O6-position of guanine by O6-alkylguanine-DNA alkyltransferase (AGT). |
|

|
INTERACTIVE GO GRAPH
|