GO TERM SUMMARY
|
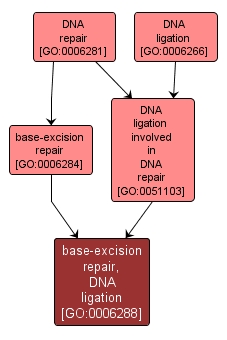
| Name: |
base-excision repair, DNA ligation |
| Acc: |
GO:0006288 |
| Aspect: |
Biological Process |
| Desc: |
The ligation by DNA ligase of DNA strands. Ligation occurs after polymerase action to fill the gap left by the action of endonucleases during base-excision repair. |
|

|
INTERACTIVE GO GRAPH
|