GO TERM SUMMARY
|
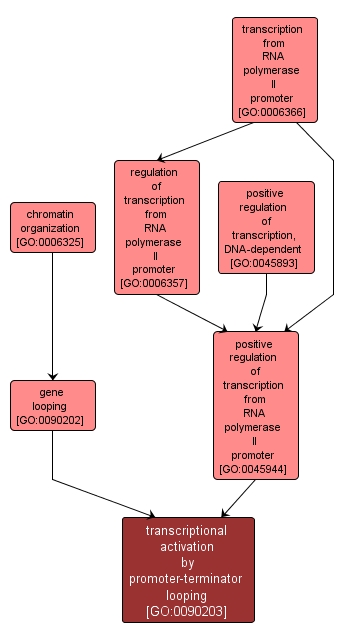
| Name: |
transcriptional activation by promoter-terminator looping |
| Acc: |
GO:0090203 |
| Aspect: |
Biological Process |
| Desc: |
The formation and maintenance of DNA loops that juxtapose the promoter and terminator regions of RNA polymerase II-transcribed genes and activate transcription from an RNA polymerase II promoter. |
Synonyms:
- transcriptional activation by memory gene loops
|
|

|
INTERACTIVE GO GRAPH
|