GO TERM SUMMARY
|
| Name: |
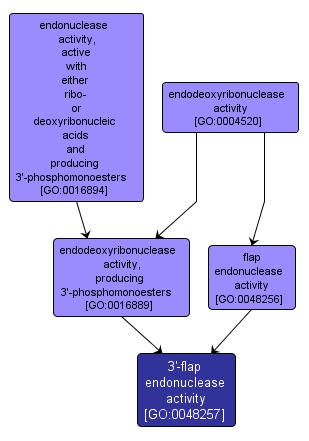
3'-flap endonuclease activity |
| Acc: |
GO:0048257 |
| Aspect: |
Molecular Function |
| Desc: |
Catalysis of the cleavage of a 3' flap structure in DNA, but not other DNA structures; processes the 3' ends of Okazaki fragments in lagging strand DNA synthesis. |
Synonyms:
- 3' flap endonuclease activity
|
|

|
INTERACTIVE GO GRAPH
|