













| General Information: |
|
| Name(s) found: |
PNG1 /
YPL096W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA mitochondrion [IDA cytosol [IDA |
| Biological Process: |
protein deglycosylation
[IDA misfolded or incompletely synthesized protein catabolic process [IMP |
| Molecular Function: |
peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity
[IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGEVYEKNNI DFDSIAKMLL IKYKDFILSK FKKAAPVENI RFQNLVHTNQ FAQGVLGQSQ 60
61 HLCTVYDNPS WHSIVLETLD LDLIYKNVDK EFAKDGHAEG ENIYTDYLVK ELLRYFKQDF 120
121 FKWCNKPDCN HCGQNTSENM TPLGSQGPNG EESKFNCGTV EIYKCNRCGN ITRFPRYNDP 180
181 IKLLETRKGR CGEWCNLFTL ILKSFGLDVR YVWNREDHVW CEYFSNFLNR WVHVDSCEQS 240
241 FDQPYIYSIN WNKKMSYCIA FGKDGVVDVS KRYILQNELP RDQIKEEDLK FLCQFITKRL 300
301 RYSLNDDEIY QLACRDEQEQ IELIRGKTQE TKSESVSAAS KSSNRGRESG SADWKAQRGE 360
361 DGK |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Riffle et al. (2010) (Unpublished Data) |

|
PNG1 RAD23 |
| View Details | Krogan NJ, et al. (2006) |

|
PNG1 RAD23 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
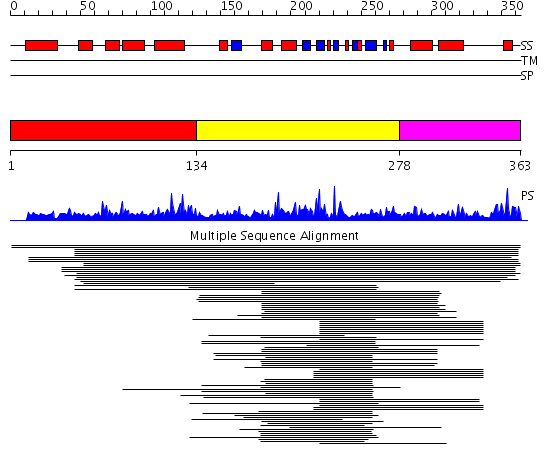
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..133] | 1.054988 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [134..277] | 15.443697 | Transglutaminase-like superfamily No confident structure predictions are available. | |
| 3 | View Details | [278..363] | 2.128995 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)