













| General Information: |
|
| Name(s) found: |
ARG7 /
YMR062C
[SGD]
|
| Description(s) found:
Found 27 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 441 amino acids |
Gene Ontology: |
|
| Cellular Component: |
mitochondrial matrix
[IDA mitochondrion [IDA |
| Biological Process: |
arginine biosynthetic process
[TAS ornithine biosynthetic process [IDA cellular cell wall organization [IMP |
| Molecular Function: |
glutamate N-acetyltransferase activity
[IDA acetyl-CoA:L-glutamate N-acetyltransferase activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRISSTLLQR SKQLIDKYAL YVPKTGSFPK GFEVGYTASG VKKNGSLDLG VILNTNKSRP 60
61 STAAAVFTTN KFKAAPVLTS KKVLETARGK NINAIVVNSG CANSVTGDLG MKDAQVMIDL 120
121 VNDKIGQKNS TLVMSTGVIG QRLQMDKIST GINKIFGEEK FGSDFNSWLN VAKSICTTDT 180
181 FPKLVTSRFK LPSGTEYTLT GMAKGAGMIC PNMATLLGFI VTDLPIESKA LQKMLTFATT 240
241 RSFNCISVDG DMSTNDTICM LANGAIDTKE INEDSKDFEQ VKLQVTEFAQ RLAQLVVRDG 300
301 EGSTKFVTVN VKNALHFEDA KIIAESISNS MLVKTALYGQ DANWGRILCA IGYAKLNDLK 360
361 SLDVNKINVS FIATDNSEPR ELKLVANGVP QLEIDETRAS EILALNDLEV SVDLGTGDQA 420
421 AQFWTCDLSH EYVTINGDYR S |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Qiu et al. (2008) |

|
ARG7 ARG8 CIT1 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
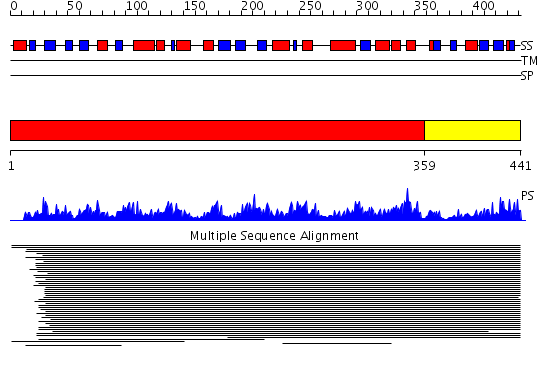
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..358] | 8.98 | L-aminopeptidase D-Ala-esterase/amidase | |
| 2 | View Details | [359..441] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)