













| General Information: |
|
| Name(s) found: |
SMP1 /
YBR182C
[SGD]
|
| Description(s) found:
Found 26 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 452 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
regulation of response to osmotic stress
[IGI positive regulation of transcription from RNA polymerase II promoter [IGI |
| Molecular Function: |
transcription factor activity
[IPI DNA bending activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGRRKIEIEP IKDDRNRTVT FIKRKAGLFK KAHELSVLCQ VDIAVIILGS NNTFYEYSSV 60
61 DMSNLLNVHQ NNTDLPHNII EPSDYGDYVK KPRVVLNERK RRRRRATVLQ PASHSGSCTV 120
121 SSQDSSSVQN NGNLSAPLAS NDAGNAGVST PLVHCHGAIS RSGSNHSDCA RNSADYQMLQ 180
181 GGLNSGGSFH ANDYKESVDQ QHVANEAIHR NFMNKRIRPD THLLLSESNH SNYHNFYPSP 240
241 YENLPKPSLP ASLVGNIPSF QSQFVQVIPA NSNPMGKGFN GTGDSESFEA KQKIHPTVAI 300
301 SNTLEGPAPV QAMVHHLHQL NSNRGKLSGK PYLKLNIPKA TNDACQRSPA MYSGTASPKT 360
361 DVQATPNQML ASNMSSPLSR SKFLGFKNND MDDLYHNGRC GSTYVNNKTF FLKPPIGRPP 420
421 KFPKSPSSSI VVFPSSVASS TLKSTSSTNS PD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
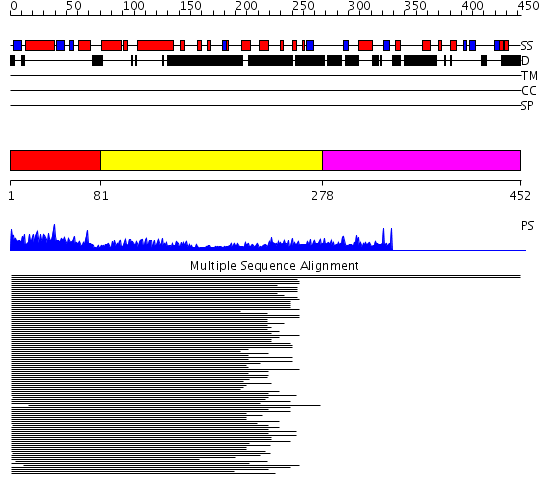
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..80] | 189.616428 | Mef2a core | |
| 2 | View Details | [81..277] | 18.490997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [278..452] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [348..452] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)