













| General Information: |
|
| Name(s) found: |
PDR3 /
YBL005W
[SGD]
|
| Description(s) found:
Found 24 descriptions. SHOW ALL |
|
| Organism: | Saccharomyces cerevisiae |
| Length: | 976 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleus
[IDA cytoplasm [IDA |
| Biological Process: |
positive regulation of transcription, DNA-dependent
[IDA response to drug [TAS regulation of transcription from RNA polymerase II promoter [TAS |
| Molecular Function: |
DNA binding
[IPI transcription activator activity [IDA |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVKKSTRSK VSTACVNCRK RKIKCTGKYP CTNCISYDCT CVFLKKHLPQ KEDSSQSLPT 60
61 TAVAPPSSHA NVEASADVQH LDTAIKLDNQ YYFKLMNDLI QTPVSPSATH APDTSNNPTN 120
121 DNNILFKDDS KYQNQLVTYQ NILTNLYALP PCDDTQLLID KTKSQLNNLI NSWNPEINYP 180
181 KLSSFSPRPQ RSIETYLLTN KYRNKIHMTR FSFWTDQMVK SQSPDSFLAT TPLVDEVFGL 240
241 FSPIQAFSLR GIGYLIKKNI ENTGSSMLID TKETIYLILR LFDLCYEHLI QGCISISNPL 300
301 ENYLQKIKQT PTTTASASLP TSPAPLSNDL VISVIHQLPQ PFIQSITGFT TTQLIENLHD 360
361 SFSMFRIVTQ MYAQHRKRFA EFLNQAFSLP HQEKSVLFSS FCSSEYLLST LCYAYYNVTL 420
421 YHMLDINTLD YLEILVSLLE IQNEIDERFG FEKMLEVAVT CSTKMGLSRW EYYVGIDENT 480
481 AERRRKIWWK IYSLEKRFLT DLGDLSLINE HQMNCLLPKD FRDMGFINHK EFLTKIGTSS 540
541 LSPSSPKLKN LSLSRLIEYG ELAIAQIVGD FFSETLYNEK FTSLEVSVKP TIIRQKLLEK 600
601 VFEDIESFRL KLAKIKLHTS RVFQVAHCKY PEYPKNDLIE AAKFVSYHKN TWFSILGAVN 660
661 NLIARLSEDP EVITEQSMKY ANEMFQEWRE INQFLIQVDT DFIVWACLDF YELIFFVMAS 720
721 KFYVEDPHIT LEDVINTLKV FKRITNIISF FNNNLDEKDY DCQTFREFSR SSSLVAISIR 780
781 IIFLKYCYAE QIDRAEFIER LKEVEPGLSD LLREFFDTRS FIYRYMLKSV EKSGFHLIIR 840
841 KMLESDYKFL YRDKLATGNI PDQGNSSQIS QLYDSTAPSY NNASASAANS PLKLSSLLNS 900
901 GEESYTQDAS ENVPCNLRHQ DRSLQQTKRQ HSAPSQISAN ENNIYNLGTL EEFVSSGDLT 960
961 DLYHTLWNDN TSYPFL |
| PUBLICATION | TOPOLOGY | COCOMPLEXED PROTEINS | |
| View Details | Krogan NJ, et al. (2006) |

|
ARA1 PDR3 |
The following runs contain data for this protein:
| BAIT | COMMENTS | PUBLICATION | |
| View Run | #03 Alpha Factor Prep1-TiO2 Flowthrough | Keck JM, et al. (2011) |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
| PROTEIN(S) | PUBLICATION | |
| View Data |
|
Huh WK, et al. (2003) |
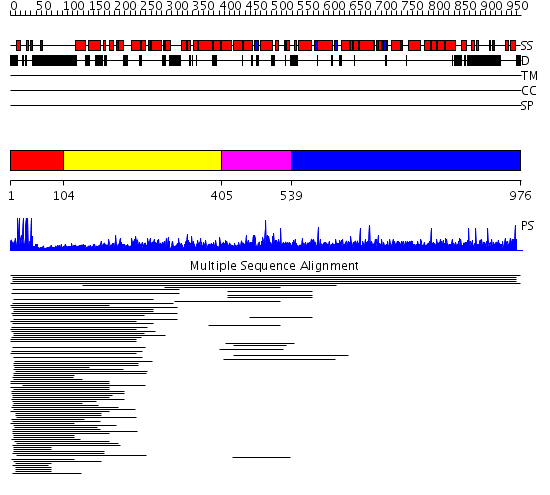
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | 46.144602 | PUT3 | |
| 2 | View Details | [104..404] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [405..538] | 9.024997 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [539..976] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [525..754] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [755..976] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)