













| General Information: |
|
| Name(s) found: |
gi|7511903
[NCBI NR]
gi|45551208 [NCBI NR] gi|45446783 [NCBI NR] gi|4165455 [NCBI NR] |
| Description(s) found:
Found 10 descriptions. SHOW ALL |
|
| Organism: | Drosophila melanogaster |
| Length: | 665 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cytoplasm
[IEA]
|
| Biological Process: |
cell redox homeostasis
[IEA]
oxidation reduction [IEA] |
| Molecular Function: |
FAD binding
[IEA]
oxidoreductase activity [IEA] 2 iron, 2 sulfur cluster binding [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDWPFCLSV SKVGGLQIRK KWQSPRTALS LVKTKRAFEG LLRIRKLPSS QSWRILNAPS 60
61 FASHINNRLL NTMGSVNCKE YKTTGSTPKK EKSTGGAVGS YQSKCSNQSP STTMSTEESS 120
121 PDSEYTSAVP VDCRVTDLKE NEMKQVDFDE DTRVLLVKQN DRLLAVGAKC THYGAPLQTG 180
181 ALGLGRVRCP WHGACFNLEN GDIEDFPGLD SLPCYRVEVG NEGQVMLRAK RSDLVNNKRL 240
241 KNMVRRKPDD QRVFIVVGGG PSGAVAVETI RQEGFTGRLI FVCREDYLPY DRVKISKAMN 300
301 LEIEQLRFRD EEFYKEYDIE LWQGVAAEKL DTAQKELHCS NGYVVKYDKI YLATGCSAFR 360
361 PPIPGVNLEN VRTVRELADT KAILASITPE SRVVCLGSSF IALEAAAGLV SKVQSVTVVG 420
421 RENVPLKAAF GAEIGQRVLQ LFEDNKVVMR MESGIAEIVG NEDGKVSEVV LVDDTRLPCD 480
481 LLILGTGSKL NTQFLAKSGV KVNRNGSVDV TDFLESNVPD VYVGGDIANA HIHGLAHDRV 540
541 NIGHYQLAQY HGRVAAINMC GGVKKLEAVP FFFTLIFGKG IRYAGHGSYK DVIIDGSMED 600
601 FKFVAYFINE ADTVTAVASC GRDPIVAQFA ELISQGKCLG RGQIEDPATR EDWTKKLGQP 660
661 LPQVR |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
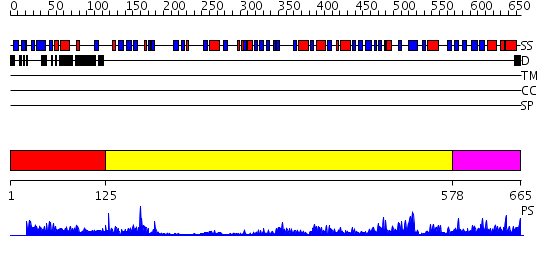
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..124] | 0.965 | Interleukin-4 receptor alpha chain | |
| 2 | View Details | [125..577] | 15.69897 | Dihydropyrimidine dehydrogenase, N-terminal domain; Dihydropyrimidine dehydrogenase, domain 4; Dihydropyrimidine dehydrogenase, domain 3; Dihydropyrimidine dehydrogenase, domain 2; Dihydropyrimidine dehydrogenase, C-terminal domain | |
| 3 | View Details | [578..665] | 64.154902 | Crystal Structure of Putidaredoxin Reductase from Pseudomonas putida |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)