













| General Information: |
|
| Name(s) found: |
YGFK_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 9 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 1032 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGDIMRPIPF EELLTRIFDE YQQQRSIFGI PEQQFYSPVK GKTVSVFGET CATPVGPAAG 60
61 PHTQLAQNIV TSWLTGGRFI ELKTVQILDR LELEKPCIDA EDECFNTEWS TEFTLLKAWD 120
121 EYLKAWFALH LLEAMFQPSD SGKSFIFNMS VGYNLEGIKQ PPMQQFIDNM MDASDHPKFA 180
181 QYRDTLNKLL QDDAFLARHG LQEKRESLQA LPARIPTSMV HGVTLSTMHG CPPHEIEAIC 240
241 RYMLEEKGLN TFVKLNPTLL GYARVREILD VCGFGYIGLK EESFDHDLKL TQALEMLERL 300
301 MALAKEKSLG FGVKLTNTLG TINNKGALPG EEMYMSGRAL FPLSINVAAV LSRAFDGKLP 360
361 ISYSGGASQL TIRDIFDTGI RPITMATDLL KPGGYLRLSA CMRELEGSDA WGLDHVDVER 420
421 LNRLAADALT MEYTQKHWKP EERIEVAEDL PLTDCYVAPC VTACAIKQDI PEYIRLLGEH 480
481 RYADALELIY QRNALPAITG HICDHQCQYN CTRLDYDSAL NIRELKKVAL EKGWDEYKQR 540
541 WHKPAGSGSR HPVAVIGAGP AGLAAGYFLA RAGHPVTLFE REANAGGVVK NIIPQFRIPA 600
601 ELIQHDIDFV AAHGVKFEYG CSPDLTIEQL KNQGFHYVLI ATGTDKNSGV KLAGDNQNVW 660
661 KSLPFLREYN KGTALKLGKH VVVVGAGNTA MDCARAALRV PGVEKATIVY RRSLQEMPAW 720
721 REEYEEALHD GVEFRFLNNP ERFDADGTLT LRVMSLGEPD EKGRRRPVET NETVTLLVDS 780
781 LITAIGEQQD TEALNAMGVP LDKNGWPDVD HNGETRLTDV FMIGDVQRGP SSIVAAVGTA 840
841 RRATDAILSR ENIRSHQNDK YWNNVNPAEI YQRKGDISIT LVNSDDRDAF VAQEAARCLE 900
901 CNYVCSKCVD VCPNRANVSI AVPGFQNRFQ TLHLDAYCNE CGNCAQFCPW NGKPYKDKIT 960
961 VFSLAQDFDN SSNPGFLVED CRVRVRLNNQ SWVLNIDSKG QFNNVPPELN DMCRIISHVH1020
1021 QHHHYLLGRV EV |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
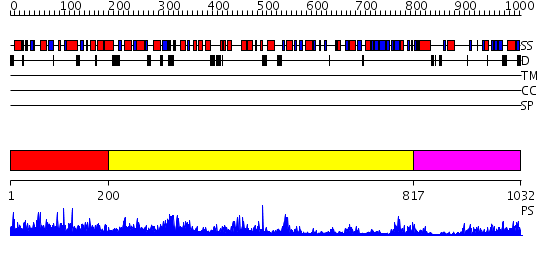
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [200..816] | 93.39794 | The Crystal Structure and Reaction Mechanism of E. coli 2,4-Dienoyl CoA Reductase | |
| 3 | View Details | [817..1032] | 65.39794 | The crystal structure of dihydrolipoamide dehydrogenase and dihydrolipoamide dehydrogenase-binding protein (didomain) subcomplex of human pyruvate dehydrogenase complex. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||
| 2 |
|
|||||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)