













| General Information: |
|
| Name(s) found: |
YDIJ_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 1018 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIPQISQAPG VVQLVLNFLQ ELEQQGFTGD TATSYADRLT MSTDNSIYQL LPDAVVFPRS 60
61 TADVALIARL AAQERYSSLI FTPRGGGTGT NGQALNQGII VDMSRHMNRI IEINPEEGWV 120
121 RVEAGVIKDQ LNQYLKPFGY FFAPELSTSN RATLGGMINT DASGQGSLVY GKTSDHVLGV 180
181 RAVLLGGDIL DTQPLPVELA ETLGKSNTTI GRIYNTVYQR CRQQRQLIID NFPKLNRFLT 240
241 GYDLRHVFND EMTEFDLTRI LTGSEGTLAF ITEARLDITR LPKVRRLVNV KYDSFDSALR 300
301 NAPFMVEARA LSVETVDSKV LNLAREDIVW HSVSELITDV PDQEMLGLNI VEFAGDDEAL 360
361 IDERVNALCA RLDELIASHQ AGVIGWQVCR ELAGVERIYA MRKKAVGLLG NAKGAAKPIP 420
421 FAEDTCVPPE HLADYIAEFR ALLDSHGLSY GMFGHVDAGV LHVRPALDMC DPQQEILMKQ 480
481 ISDDVVALTA KYGGLLWGEH GKGFRAEYSP AFFGEELFAE LRKVKAAFDP HNRLNPGKIC 540
541 PPEGLDAPMM KVDAVKRGTF DRQIPIAVRQ QWRGAMECNG NGLCFNFDAR SPMCPSMKIT 600
601 QNRIHSPKGR ATLVREWLRL LADRGVDPLK LEQELPESGV SLRTLIARTR NSWHANKGEY 660
661 DFSHEVKEAM SGCLACKACS TQCPIKIDVP EFRSRFLQLY HTRYLRPLRD HLVATVESYA 720
721 PLMARAPKTF NFFINQPLVR KLSEKHIGMV DLPLLSVPSL QQQMVGHRSA NMTLEQLESL 780
781 NAEQKARTVL VVQDPFTSYY DAQVVADFVR LVEKLGFQPV LLPFSPNGKA QHIKGFLNRF 840
841 AKTAKKTADF LNRMAKLGMP MVGVDPALVL CYRDEYKLAL GEERGEFNVL LANEWLASAL 900
901 ESQPVATVSG ESWYFFGHCT EVTALPGAPA QWAAIFARFG AKLENVSVGC CGMAGTYGHE 960
961 AKNHENSLGI YELSWHQAMQ RLPRNRCLAT GYSCRSQVKR VEGTGVRHPV QALLEIIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
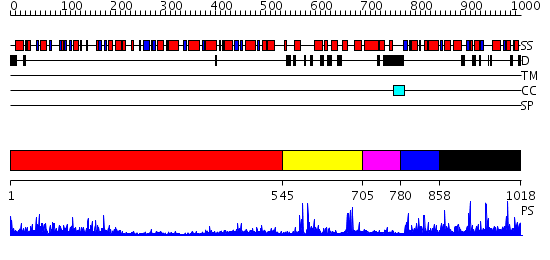
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..544] | 49.0 | No description for 2uuuA was found. | |
| 2 | View Details | [545..704] | 4.69897 | Fumarate reductase; Fumarate reductase iron-sulfur protein, N-terminal domain | |
| 3 | View Details | [705..779] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [780..857] | 2.452996 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [858..1018] | 7.452993 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)