













| General Information: |
|
| Name(s) found: |
SYV_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 7 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 951 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MEKTYNPQDI EQPLYEHWEK QGYFKPNGDE SQESFCIMIP PPNVTGSLHM GHAFQQTIMD 60
61 TMIRYQRMQG KNTLWQVGTD HAGIATQMVV ERKIAAEEGK TRHDYGREAF IDKIWEWKAE 120
121 SGGTITRQMR RLGNSVDWER ERFTMDEGLS NAVKEVFVRL YKEDLIYRGK RLVNWDPKLR 180
181 TAISDLEVEN RESKGSMWHI RYPLADGAKT ADGKDYLVVA TTRPETLLGD TGVAVNPEDP 240
241 RYKDLIGKYV ILPLVNRRIP IVGDEHADME KGTGCVKITP AHDFNDYEVG KRHALPMINI 300
301 LTFDGDIRES AQVFDTKGNE SDVYSSEIPA EFQKLERFAA RKAVVAAVDA LGLLEEIKPH 360
361 DLTVPYGDRG GVVIEPMLTD QWYVRADVLA KPAVEAVENG DIQFVPKQYE NMYFSWMRDI 420
421 QDWCISRQLW WGHRIPAWYD EAGNVYVGRN EDEVRKENNL GADVVLRQDE DVLDTWFSSA 480
481 LWTFSTLGWP ENTDALRQFH PTSVMVSGFD IIFFWIARMI MMTMHFIKDE NGKPQVPFHT 540
541 VYMTGLIRDD EGQKMSKSKG NVIDPLDMVD GISLPELLEK RTGNMMQPQL ADKIRKRTEK 600
601 QFPNGIEPHG TDALRFTLAA LASTGRDINW DMKRLEGYRN FCNKLWNASR FVLMNTEGQD 660
661 CGFNGGEMTL SLADRWILAE FNQTIKAYRE ALDSFRFDIA AGILYEFTWN QFCDWYLELT 720
721 KPVMNGGTEA ELRGTRHTLV TVLEGLLRLA HPIIPFITET IWQRVKVLCG ITADTIMLQP 780
781 FPQYDASQVD EAALADTEWL KQAIVAVRNI RAEMNIAPGK PLELLLRGCS ADAERRVNEN 840
841 RGFLQTLARL ESITVLPADD KGPVSVTKII DGAELLIPMA GLINKEDELA RLAKEVAKIE 900
901 GEISRIENKL ANEGFVARAP EAVIAKEREK LEGYAEAKAK LIEQQAVIAA L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
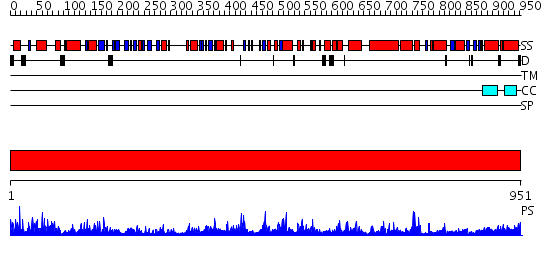
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..951] | 1000.0 | Valyl-tRNA synthetase (ValRS) C-terminal domain; Valyl-tRNA synthetase (ValRS) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)