













| General Information: |
|
| Name(s) found: |
gi|151942308
[NCBI NR]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae YJM789 |
| Length: | 689 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTSTLATRLS TYSISLILQR IKIIKRCYSA PVLRDYQQDA IDACVNSIRQ GTKRIGVSLA 60
61 TGGGKTVIFS NLINQLRQNY FKERQGNFKS LILVHRRELA LQATATLKKI FPDLKVHIEM 120
121 GKYDCDIEDS DVIVASVQTL IRRLHKYDTN SVNLIIIDEA HHSVANSYRS ILDHFKASTA 180
181 ETKIPVIGFS ATFERADKRA LSMVMDKIVY HRGILEMIDD KWLCEAKFTS VKIEADLSDV 240
241 KSTADDFQLA PLSSLMNTKE INEVILKTYL HKKQEKSLKS TLLFGVDKAH VQSLHKLFKD 300
301 NGINTDYVTS DTKQIERDNI IQKFKNGETE VLMNCGIFTE GTDMPNIDCI LLCRPTKSRS 360
361 LLIQMIGRGL RLHHSKDHCH IIDFIGASSV GVVSAPTLLG IRSDDIEFDD ATVEDLKAIQ 420
421 GEIIAKQQKI DERLRALFQT DEAAMENVTE RNSVADWIHS ANSVDLTLCS FDSFRNFTQS 480
481 NNSYPSGKEF DEASEAVKEM ELLMNSQYPW VKFASNAWGL PLKGKNHLRI YKEKSEDKLS 540
541 MVYHLKMYRQ LPCFITNKYA DYVPKSIIKD ANLWNVMSKV EKIINTLNSD LEGQTMQYQA 600
601 ISSKYSKWRQ TVPTSKQRDF VFRKLKKVYG ESSKDFIRLS LDDVTTYVNT KMTKGDASNL 660
661 IFASSLAPVY PLKSLLRILE YQKRRSFIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
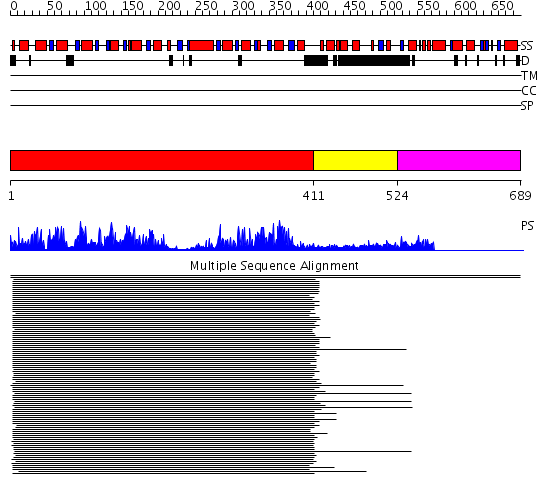
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..410] | 132.228787 | Initiation factor 4a | |
| 2 | View Details | [411..523] | 72.228787 | Nucleotide excision repair enzyme UvrB | |
| 3 | View Details | [524..689] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||
| 3 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.92 |
Source: Reynolds et al. (2008)