













| General Information: |
|
| Name(s) found: |
FLHA_ECOLI
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Escherichia coli |
| Length: | 692 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNLAAMLRL PANLKSTQWQ ILAGPILILL ILSMMVLPLP AFILDLLFTF NIALSIMVLL 60
61 VAMFTQRTLE FAAFPTILLF TTLLRLALNV ASTRIILMEG HTGAAAAGKV VEAFGHFLVG 120
121 GNFAIGIVVF VILVIINFMV ITKGAGRIAE VGARFVLDGM PGKQMAIDAD LNAGLIGEDE 180
181 AKKRRSEVTQ EADFYGSMDG ASKFVRGDAI AGILIMVINI VGGLLVGVLQ HGMSMGHAAE 240
241 SYTLLTIGDG LVAQIPALVI STAAGVIVTR VSTDQDVGEQ MVNQLFSNPS VMLLSAAVLG 300
301 LLGLVPGMPN LVFLLFTAGL LGLAWWIRGR EQKAPAEPKP VKMAENNTVV EATWNDVQLE 360
361 DSLGMEVGYR LIPMVDFQQD GELLGRIRSI RKKFAQEMGF LPPVVHIRDN MDLQPARYRI 420
421 LMKGVEIGSG DAYPGRWLAI NPGTAAGTLP GEATVDPAFG LNAIWIESAL KEQAQIQGYT 480
481 VVEASTVVAT HLNHLISQHA AELFGRQEAQ QLLDRVAQEM PKLTEDLVPG VVTLTTLHKV 540
541 LQNLLDEKVP IRDMRTILET LAEHAPIQSD PHELTAVVRV ALGRAITQQW FPGKDEVHVI 600
601 GLDTPLERLL LQALQGGGGL EPGLADRLLA QTQEALSRQE MLGAPPVLLV NHALRPLLSR 660
661 FLRRSLPQLV VLSNLELSDN RHIRMTATIG GK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
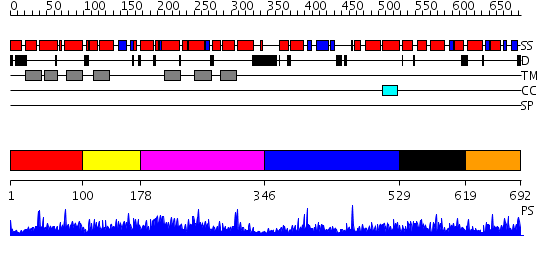
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..99] | 6.152994 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [100..177] | 2.163987 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [178..345] | 1.163978 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [346..528] | 2.163986 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [529..618] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [619..692] | 4.155995 | View MSA. No confident structure predictions are available. |
| Source: Reynolds et al. 2008. Manuscript submitted |

|