













| General Information: |
|
| Name(s) found: |
GLO4_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 363 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
metabolic process
[IEA]
|
| Molecular Function: |
FMN binding
[IEA]
glycolate oxidase activity [ISS] catalytic activity [IEA] oxidoreductase activity [IEA] electron carrier activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDQIVNVDEF QELAKQALPK MYYDFYNGGA EDQHTLNENV QAFRRIMFRP RVLVDVSNID 60
61 MSTSMLGYPI SAPIMIAPTA MHKLAHPKGE IATAKAAAAC NTIMIVSFMS TCTIEEVASS 120
121 CNAVRFLQIY VYKRRDVTAQ IVKRAEKAGF KAIVLTVDVP RLGRREADIK NKMISPQLKN 180
181 FEGLVSTEVR PNEGSGVEAF ASSAFDASLS WKDIEWLRSI TKLPILVKGL LTREDALKAV 240
241 EAGVDGIVVS NHGARQLDYS PATITVLEEV VHAVKGRIPV LLDGGVRRGT DVFKALALGA 300
301 QAVLIGRPIV YGLAAKGEDG VKKVIDMLKN EFEITMALSG CPTIDDVTRN HVRTENERIK 360
361 SML |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
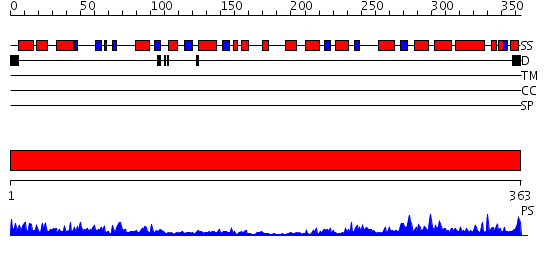
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..363] | 87.522879 | THREE-DIMENSIONAL STRUCTURES OF GLYCOLATE OXIDASE WITH BOUND ACTIVE-SITE INHIBITORS |
Functions predicted (by domain):
| # | Gene Ontology predictions |
| 1 |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.85 |
Source: Reynolds et al. (2008)