













| General Information: |
|
| Name(s) found: |
SSY2_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 13 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 792 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
|
| Biological Process: |
glucan biosynthetic process
[IEA]
biosynthetic process [IEA] |
| Molecular Function: |
transferase activity, transferring glycosyl groups
[ISS]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVAESSFP LLCQIKTQRR INSSTLRHSR VSYHDLPSGS LSFRSRSFVL GHRCKCVSRV 60
61 EASGSDDDEP EDALQATIDK SKKVLAMQRN LLHQIAERRK LVSSIKESTP DLDDAKASSK 120
121 QESASSVNAN TDATKKEIMD GDANGSVSPS TYGKSSLSKE PEAKTFSPST ESLKNRKQSS 180
181 ASVISSSPVT SPQKPSDVAT NGKPWSSVVA SSVDPPYKPS SVMTSPEKTS DPVTSPGKPS 240
241 KSRAGAFWSD PLPSYLTKAP QTSTMKTEKY VEKTPDVASS ETNEPGKDEE KPPPLAGANV 300
301 MNVILVAAEC APFSKTGGLG DVAGALPKSL ARRGHRVMVV VPRYAEYAEA KDLGVRKRYK 360
361 VAGQDMEVMY FHAFIDGVDF VFIDSPEFRH LSNNIYGGNR LDILKRMVLF CKAAVEVPWY 420
421 VPCGGVCYGD GNLAFIANDW HTALLPVYLK AYYRDHGIMK YTRSVLVIHN IAHQGRGPVD 480
481 DFSYVDLPSH YLDSFKLYDP VGGEHFNIFA AGLKAADRVL TVSHGYSWEV KTLEGGWGLH 540
541 NIINENDWKF RGIVNGIDTQ EWNPEFDTYL HSDDYTNYSL ENLHIGKPQC KAALQKELGL 600
601 PVRPDVPLIG FIGRLDHQKG VDLIAEAVPW MMSQDVQLVM LGTGRPDLEE VLRQMEHQYR 660
661 DKARGWVGFS VKTAHRITAG ADILLMPSRF EPCGLNQLYA MNYGTIPVVH AVGGLRDTVQ 720
721 QFDPYSETGL GWTFDSAEAG KLIHALGNCL LTYREYKESW EGLQRRGMTQ DLSWDNAAEK 780
781 YEEVLVAAKY HW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
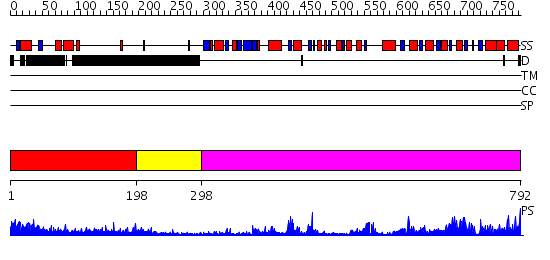
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..197] | 28.0 | RBP1 | |
| 2 | View Details | [198..297] | 1.51 | No description for 2j63A was found. | |
| 3 | View Details | [298..792] | 88.39794 | Crystal structure of the glycogen synthase from A. tumefaciens in complex with ADP |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 3 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)