













| General Information: |
|
| Name(s) found: |
HSP7H_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 16 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 563 amino acids |
Gene Ontology: |
|
| Cellular Component: |
cellular_component
[ND]
|
| Biological Process: |
response to heat
[IEP]
response to high light intensity [IEP] response to hydrogen peroxide [IEP] protein folding [TAS] |
| Molecular Function: |
ATP binding
[IEA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAEAAYTVAS DSENTGEEKS SSSPSLPEIA LGIDIGTSQC SIAVWNGSQV HILRNTRNQK 60
61 LIKSFVTFKD EVPAGGVSNQ LAHEQEMLTG AAIFNMKRLV GRVDTDPVVH ASKNLPFLVQ 120
121 TLDIGVRPFI AALVNNAWRS TTPEEVLAIF LVELRLMAEA QLKRPVRNVV LTVPVSFSRF 180
181 QLTRFERACA MAGLHVLRLM PEPTAIALLY AQQQQMTTHD NMGSGSERLA VIFNMGAGYC 240
241 DVAVTATAGG VSQIKALAGS PIGGEDILQN TIRHIAPPNE EASGLLRVAA QDAIHRLTDQ 300
301 ENVQIEVDLG NGNKISKVLD RLEFEEVNQK VFEECERLVV QCLRDARVNG GDIDDLIMVG 360
361 GCSYIPKVRT IIKNVCKKDE IYKGVNPLEA AVRGAALEGA VTSGIHDPFG SLDLLTIQAT 420
421 PLAVGVRANG NKFIPVIPRN TMVPARKDLF FTTVQDNQKE ALIIIYEGEG ETVEENHLLG 480
481 YFKLVGIPPA PKGVPEINVC MDIDASNALR VFAAVLMPGS SSPVVPVIEV RMPTVDDGHG 540
541 WCAQALNVKY GATLDLITLQ RKM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
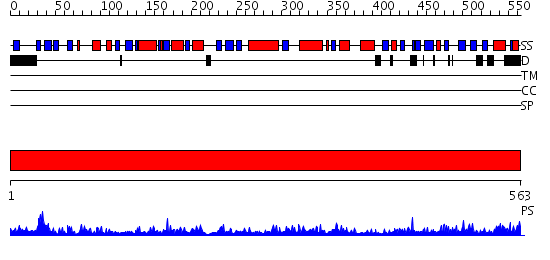
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..563] | 174.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.94 |
Source: Reynolds et al. (2008)