













| General Information: |
|
| Name(s) found: |
RH32_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 12 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 739 amino acids |
Gene Ontology: |
|
| Cellular Component: |
nucleolus
[IDA]
|
| Biological Process: |
biological_process
[ND]
|
| Molecular Function: |
nucleic acid binding
[IEA]
ATP binding [IEA] ATP-dependent helicase activity [IEA][ISS] helicase activity [IEA] |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVKIKKTKGM RKQIRLNEVE EINLLKQWIE SQKPDSGFNP LSLRPLPKDS KIGKSEDGKN 60
61 GTVFSRYAGV RKFAQLPISD KTKRGLKDAK YVDMTDVQSA AIPHALCGRD ILGAARTGSG 120
121 KTLAFVIPIL EKLHRERWSP EDGVGCIIIS PTRELAAQTF GVLNKVGKFH KFSAGLLIGG 180
181 REGVDVEKER VHEMNILVCA PGRLLQHMDE TPNFECPQLQ ILILDEADRV LDSAFKGQLD 240
241 PIISQLPKHR QTLLFSATQT KKVKDLARLS LRDPEYISVH AEAVTATPTS LMQTVMIVPV 300
301 EKKLDMLWSF IKTHLNSRIL VFLSTKKQVK FVHEAFNKLR PGIPLKSLHG KMSQEKRMGV 360
361 YSQFIERQSV LFCTDVLARG LDFDKAVDWV VQVDCPEDVA SYIHRVGRTA RFYTQGKSLL 420
421 FLTPSEEKMI EKLQEAKVPI KLIKANNQKL QEVSRLLAAL LVKYPDLQGV AQRAFITYLR 480
481 SIHKRRDKEI FDVSKLSIEN FSASLGLPMT PRIRFTNLKT KKKGVYESSI AMEIENAQEY 540
541 EAPLVVKKDL LGEDLEEEDF ALKPRKEGKV VEKSTKEEEV LIPGNRVLKN KKLKINLHRP 600
601 FGSRVVLDEE GNSLAPLASV AAEAGTEVAL DEERMNDYYK KVGAEMRKAD IEDKKVDKER 660
661 RREKRMKQKI KRKRGAMEDE EEEEEEDHDG SGSSDDETGR NSKRAKKIVS DNEENGGKIN 720
721 TDSLSVADLE EMALKFITQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
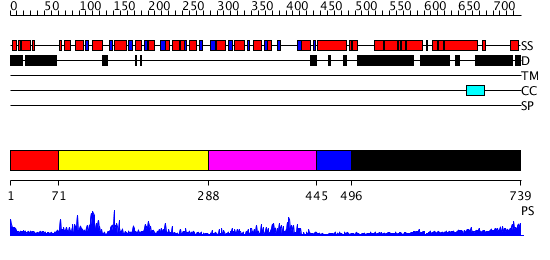
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..70] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [71..287] | 49.69897 | No description for 2pl3A was found. | |
| 3 | View Details | [288..444] | 27.221849 | No description for 2hjvA was found. | |
| 4 | View Details | [445..495] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [496..739] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)