













| General Information: |
|
| Name(s) found: |
HSP7J_ARATH
[Swiss-Prot]
|
| Description(s) found:
Found 14 descriptions. SHOW ALL |
|
| Organism: | Arabidopsis thaliana |
| Length: | 682 amino acids |
Gene Ontology: |
|
| Cellular Component: |
chloroplast
[IDA]
mitochondrial matrix [TAS] mitochondrion [IDA] cell wall [IDA] plasma membrane [IDA] |
| Biological Process: |
response to heat
[IEP]
response to salt stress [IEP] response to cadmium ion [IEP] protein folding [TAS] response to virus [IEP] |
| Molecular Function: |
ATP binding
[IDA]
|
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MATAALLRSI RRREVVSSPF SAYRCLSSSG KASLNSSYLG QNFRSFSRAF SSKPAGNDVI 60
61 GIDLGTTNSC VAVMEGKNPK VIENAEGART TPSVVAFNTK GELLVGTPAK RQAVTNPTNT 120
121 VSGTKRLIGR KFDDPQTQKE MKMVPYKIVR APNGDAWVEA NGQQYSPSQI GAFILTKMKE 180
181 TAEAYLGKSV TKAVVTVPAY FNDAQRQATK DAGRIAGLDV ERIINEPTAA ALSYGMTNKE 240
241 GLIAVFDLGG GTFDVSVLEI SNGVFEVKAT NGDTFLGGED FDNALLDFLV NEFKTTEGID 300
301 LAKDRLALQR LREAAEKAKI ELSSTSQTEI NLPFITADAS GAKHFNITLT RSRFETLVNH 360
361 LIERTRDPCK NCLKDAGISA KEVDEVLLVG GMTRVPKVQS IVAEIFGKSP SKGVNPDEAV 420
421 AMGAALQGGI LRGDVKELLL LDVTPLSLGI ETLGGVFTRL ITRNTTIPTK KSQVFSTAAD 480
481 NQTQVGIRVL QGEREMATDN KLLGEFDLVG IPPSPRGVPQ IEVTFDIDAN GIVTVSAKDK 540
541 TTGKVQQITI RSSGGLSEDD IQKMVREAEL HAQKDKERKE LIDTKNTADT TIYSIEKSLG 600
601 EYREKIPSEI AKEIEDAVAD LRSASSGDDL NEIKAKIEAA NKAVSKIGEH MSGGSGGGSA 660
661 PGGGSEGGSD QAPEAEYEEV KK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
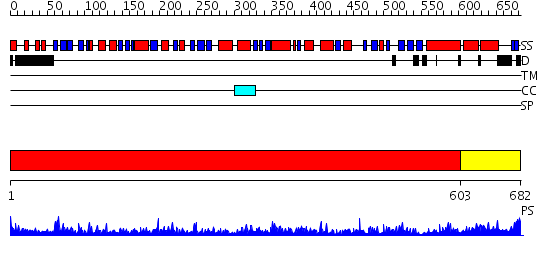
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..602] | 174.0 | crystal structure of bovine hsc70(aa1-554)E213A/D214A mutant | |
| 2 | View Details | [603..682] | 56.69897 | DnaK |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.66 |
Source: Reynolds et al. (2008)